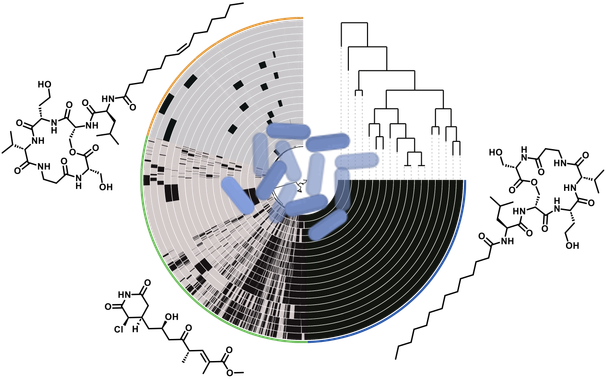
The plant pathogen #Pseudomonas syringae adapts to its environment using unique #NaturalProducts. We identified new compounds that help the bacterium to fight competitors and thrive in diverse ecological niches.
🗞️ PR: https://lmy.de/vcZyU
📝 Publication in Angewandte Chemie International Edition: https://doi.org/10.1002/anie.202503679
#NewPaper #PlantPathogens #Syrilipamides #Secimides #MicrobialDefense #BacterialGenomics #GenomicAnalysis #GeneCluster #Metabolomics #LeibnizHKI