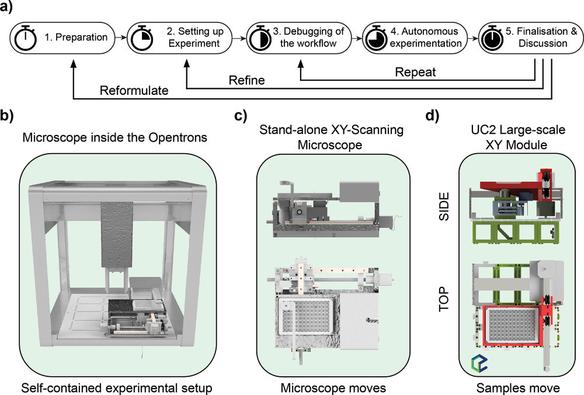
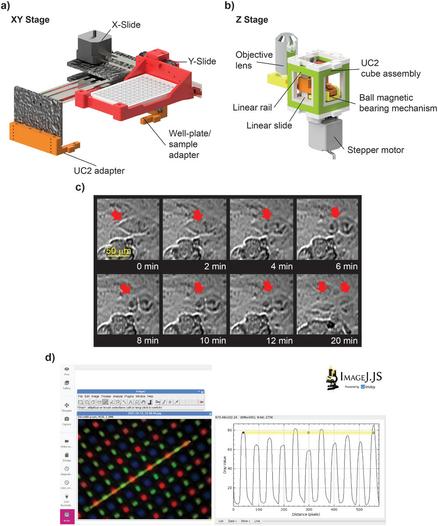
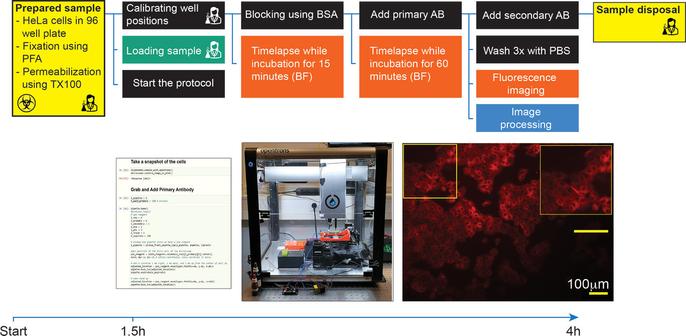
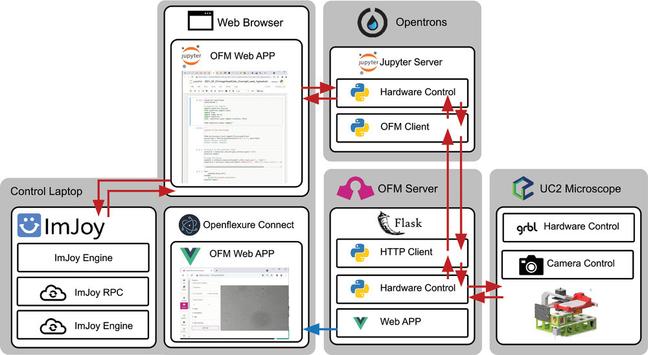
We have a new preprint and GitHub page up describing OT-Mation, a script to move experimental arrays generated using statistical Design of Experiments (#DoE) directly in to code to control the OpenTrons OT-2 liquid handling robotics platform. This reduces programming time for the OT-2 and helps prevent inclusion of incorrect experimental combinations in the script.
preprint on biorxiv:
https://www.biorxiv.org/content/10.1101/2025.01.20.633898v1