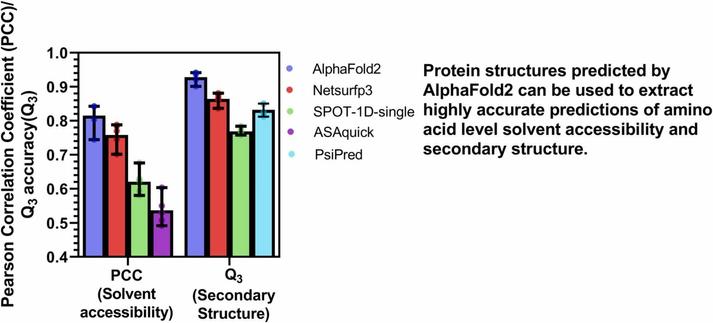
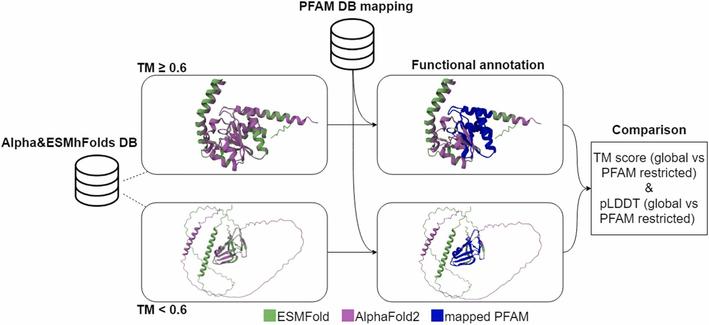
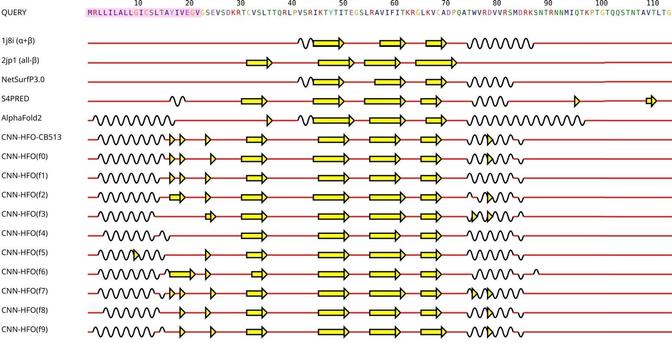
🧩 What happens when we put AlphaFold2’s predictions to the ultimate test—across species and structure types?
🔗 Comprehensive assessment of AlphaFold’s predictions of secondary structure and solvent accessibility at the amino acid-level in eukaryotic, bacterial and archaeal proteins. Computational and Structural Biotechnology Journal, DOI: https://doi.org/10.1016/j.csbj.2025.05.047
📚 CSBJ: https://www.csbj.org/
#AlphaFold #Bioinformatics #StructuralBiology #AlphaFold2 #ProteinStructure #ProteinPrediction #AI