Here's a link to the pre-print again https://arxiv.org/abs/2505.18470, and many thanks to all the others involved, Janna Hastings, @justaddcoffee, Daniel Korn and special thanks to the ChEBI team Adnan Malik and Noel O'Boyle for checking the results. May the force be with you!
Berkeley Lab, I work on #GeneOntology #MonarchInitiative #AllianceGenome #NationalMicrobimeDataCollaborative #OBOFoundry.
We have more of our findings up on a website here: https://chemkg.github.io/c3p, along with the code we used to learn the classes.
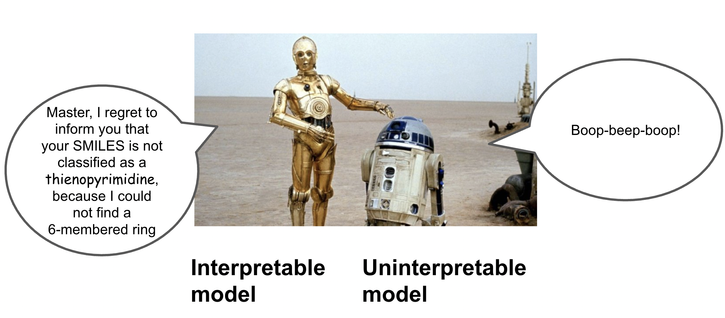
One thing to note is that even though we used LLMs to learn the classes, the resulting programs can be executed for classification with no other runtime dependencies beyond python and rdkit, and the results are fully interpretable!
additionally we suspected that there are a number of misclassifications in ChEBI that end up diverting the learning process. We used a combination of LLM vision models (viewing the chemical structures) and literature search to confirm some of these cases, and then after validation by a ChEBI curator, these were fixed in the ontology.
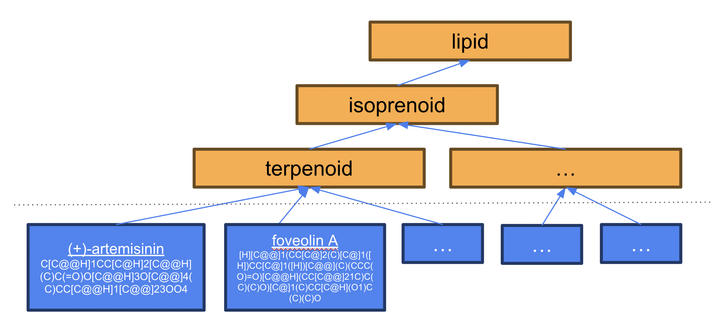
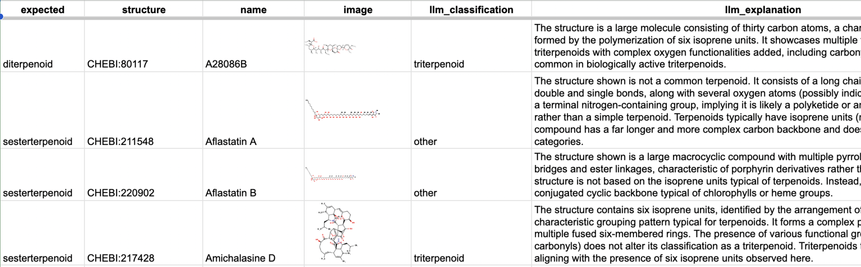
We investigated the cases where we expected the LLM to do better than it did. For example, terpenoids should in theory be easily classifiable by counting the number of isoprene units. But it turns out not to be that straightforward, chemicals can be classified both by origin or by structures....
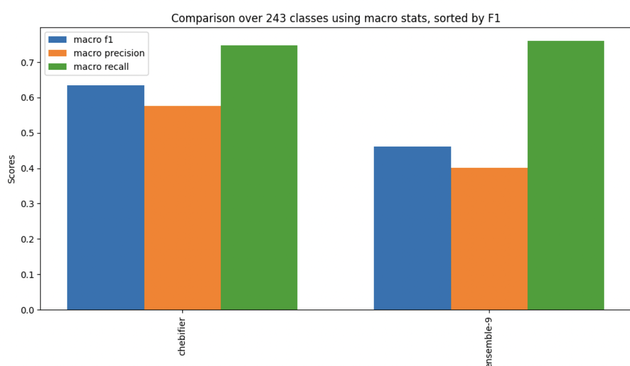
Overall the results were promising, although we fell short of the current state of the art for automated classification, chebifier.
The agent’s decision process was recorded via GitHub commits so you can look at the evolution of each individual program, along with the LLM’s “thinking process”. Here is glycerophosphocholine, which eventually converged on an f1 of 0.97
https://github.com/chemkg/c3p/commits/main/c3p/programs/glycerophosphocholine.py
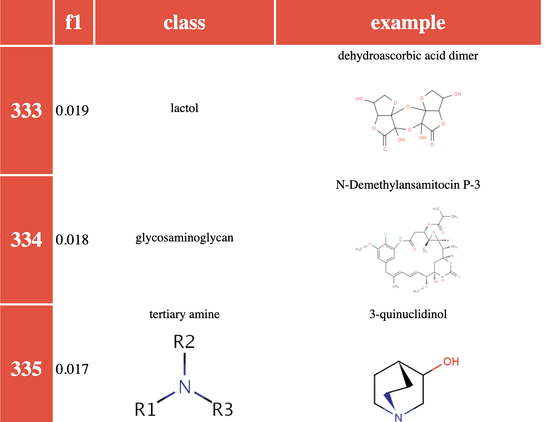
But some classes just couldn’t be learned well – and some of these were surprising.
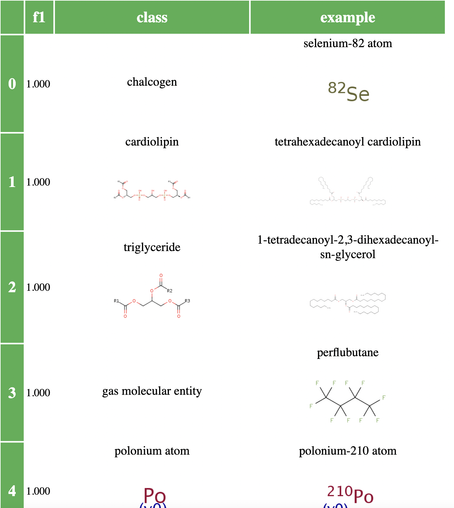
We did an experiment where we tried this on a subset of ChEBI classes (we excluded the more obscure ones, focusing on those of most relevance to biologists). It turns out that there are a lot of chemical classes that can easily be learned (well defined structures like triglycerides, as well as trivial classes for things like polonium atoms – we didn’t filter non polyatomic classes!).
But there is no open library of programs (“ontology”) for all major chemical classes, and writing this would take some time. We thought, what if we use LLMs to generate a program for each class? We could benchmark the results against existing curated classifications in CHEBI and iteratively improve them, eventually building up a ChEBI Chemical Classification Programs Ontology (C3PO). This could be used to classify new structures added to ChEBI - or any structures.
Unlike some other domains where ontologies are used, classification of chemical structures is in theory relatively crisp and deterministic, and we should be able to write python programs using libraries like RDKit that can accurately classify a SMILES string based on objective features like number of rings, counting atoms and so on.
This achieves high accuracy, but the underlying embeddings are hard for humans to introspect or tweak the classifications. Instead of learning latent representations, what if we instead learn interpretable rules in the form of programs?
Deep learning approaches to classification typically learn a latent representation of the inputs and use this to predict classes. For chemical classification, the leading deep learning approach is the awesome Chebifier, which works off of embeddings of SMILES strings. See https://chebifier.hastingslab.org/ and Glauer et al https://pubs.rsc.org/en/content/articlelanding/2024/dd/d3dd00238a
How can we scale up manual classification of chemical structures in databases like ChEBI? Can we help curators place new structures into classes like "terpenoid", based on their chemical structure? We describe a new approach in our manuscript "Chemical classification program synthesis using generative artificial intelligence" (aka the “C3PO project), pre-print here: https://arxiv.org/abs/2505.18470
@jerven Done, and thanks for your contributions, these are out in a new release: https://github.com/chemkg/chemrof/releases/tag/v0.3.0
@jerven answered your issue in GitHub, thanks for the prompt!
@jerven @cthoyt but many people just need the properties (https://chemkg.github.io/chemrof/#slots), and ChEBI themselves are using some of the properties for the new ChEBI, we are using these for our materials KG at my institute, this is where a lot of current efforts are...
@jerven @cthoyt Using it to create our benchmarks for our AI-driven chemical classification program synthesis https://chemkg.github.io/c3p/
@jerven @cthoyt more database-style ontologies need this explicit level of metaclasses to manage the different levels in the hierarchy (see https://github.com/OBOFoundry/OBOFoundry.github.io/issues/2454) -- but this is especially the case for ChEBI
@jerven @cthoyt chemrof provides the framework for which the different levels of the hierarchy should be managed https://chemkg.github.io/chemrof/ontology/
@jerven @cthoyt ChEBI desperately needs to be simplified and move towards a modern system for managing the classes, this is agreed on after the last ChEBI workshop and the new group lead is on board, slides here: Mungall, C. (2024, November 19). The need for a simpler collaboratively maintained CHEBI hierarchy. CHEBI 2024 Workshop, Hinxton, UK. Zenodo. https://doi.org/10.5281/zenodo.14298221
(O)OCC[N+](C)(C)C"
# Pattern 2: lysophosphatidylcholine with one acyl chain at position sn-1.
pattern_lyso_sn1 = "OC(=O)OCC(O)CO[P](=O)(O)OCC[N+](C)(C)C"
# Pattern 3: lysophosphatidylcholine with one acyl chain at position sn-2.
# In the SMARTS below, the '*' after OC(=O) allows for any carbon chain.
pattern_lyso_sn2 = "OCC(OC(=O)*)CO[P](=O)(O)OCC[N+](C)(C)C"](https://files.mastodon.social/cache/media_attachments/files/114/583/027/239/766/050/small/b0347762612dfb9f.png)