Adding to this thread, Yunlong Cao has posted the first experimental data on neutralization escape by BA.2.86: https://twitter.com/yunlong_cao/status/1697318194976010446
Lab studying molecular evolution of proteins and viruses. Affiliated with Fred Hutch and HHMI.
More broadly, please interactively explore the new deep mutational scanning data at https://dms-vep.github.io/SARS-CoV-2_XBB.1.5_spike_DMS/htmls/summary_overlaid.html
We hope to post full pre-print related to these data within next month.
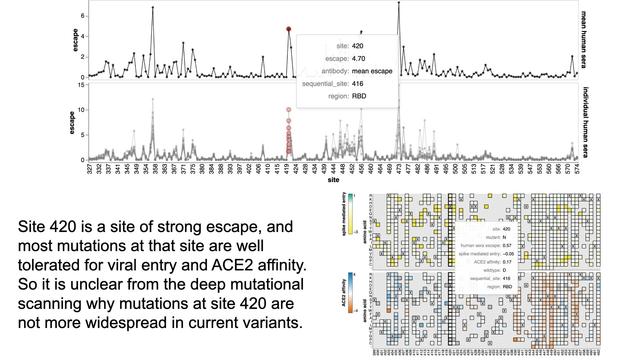
Mutations at site 420 cause appreciable escape, and also are well tolerated in deep mutational scanning. So not sure why such mutations not more widespread in nature? I guess a reminder that deep mutational scanning isn’t measuring everything that shapes real evolution!
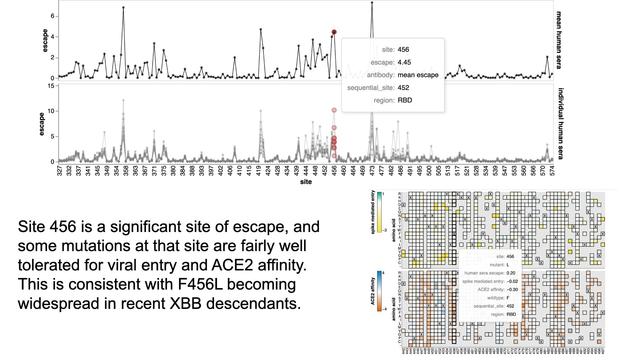
Mutations at site 456 cause appreciable escape, and are reasonably tolerated for ACE2 affinity. Likely why F456L is becoming widespread in XBB descendants. Note @yunlong_cao & David Ho have both already shown F456L causes ~2-fold drop in XBB neutralization.
Mutations at site 357 also cause substantial escape. However, most (but not all) mutations at this site are at least moderately bad for ACE2 affinity. Note that mutations like K356T (which add N-linked glycan at site 354) also may escape some of these antibodies.
Going through these top escape sites:
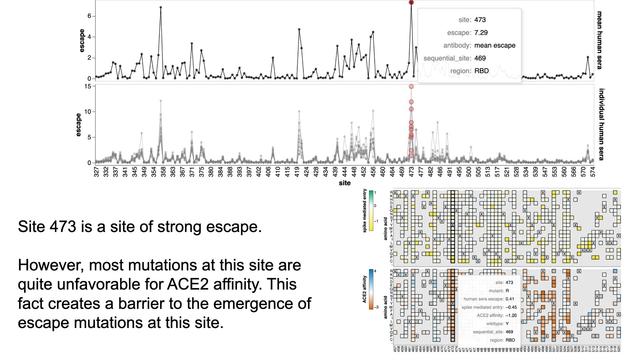
Mutations at site 473 cause substantial decrease in neutralization by XBB breakthrough sera. However, mutations at this site are also fairly bad for ACE2 affinity, creating a high barrier for such mutations to spread.
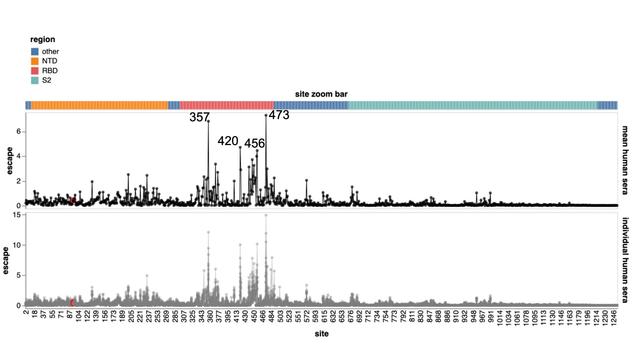
Experiments map sites in XBB spike where mutations escape neutralization by sera of humans w recent XBB infection
(Antibody response depends on exposure history, so escape may differ for other exposures not studied here)
Greatest escape is at sites 473, 357, 455/456, 420
Here are data from new XBB.1.5 spike deep mutational scanning by Bernadeta Dadonaite in our group
Go to https://dms-vep.github.io/SARS-CoV-2_XBB.1.5_spike_DMS/htmls/summary_overlaid.html to see how mutations affect:
1⃣ Neutralization by human XBB breakthrough sera
2⃣ Spike-mediated pseudovirus entry in 293T-ACE2 cells
3⃣ Affinity for ACE2
It will be few weeks before we finish pre-print for study, but we wanted to share data now for those interested in SARS2 evolution
Measurements made using lentiviral deep mutational scanning: https://sciencedirect.com/science/article/pii/S0092867423001034
I have updated my BA.2.86 slides: https://slides.com/jbloom/new_2nd_gen_ba2_variant
Slides are still up-to-date with regards to spike protein mutations.
But enough BA.2.86 sequences have now identified that slides are no longer an effective way to track those.
Instead see this tree: https://nextstrain.org/groups/neherlab/ncov/BA.2.86
Also the results of wastewater sequencing: https://twitter.com/dr_leshan/status/1694368402624893045
https://twitter.com/TanjaStadler_CH/status/1694298380841996613
I have updated my slides to reflect identification of 7th sequence of highly mutated BA.2.86 lineage: https://slides.com/jbloom/new_2nd_gen_ba2_variant
For latest on details of BA.2.86 sequences, see this Nextstrain tree from @corneliusroemer @richardneher: https://twitter.com/CorneliusRoemer/status/1693957492399599765
I have slightly updated slides referenced in this thread again (https://slides.com/jbloom/new_2nd_gen_ba2_variant) to include the England sequence recently identified as the fifth representative of the highly mutation BA.2.86 lineage.
I have slightly updated the slides referenced in this thread (https://slides.com/jbloom/new_2nd_gen_ba2_variant) to include the Michigan sequence recently identified as the fourth representative of the highly mutated BA.2.86 lineage.
https://twitter.com/SolidEvidence/status/1692200994632089809
Of course, to spread widely, antigenic advantage would need to be combined w inherent transmissibility close to best XBB variants
With only three sequences so far, there isn’t currently evidence that is case
But important to monitor if more sequences of new variant appear.
In fact, antigenic advantage of new variant over XBB is likely larger than indicated above, as many people have recently had XBB-specific antibodies boosted by infection. New variant differs from XBB at many key antigenic sites (see https://slides.com/jbloom/new_2nd_gen_ba2_variant for details)
To assess overall antigenic of mutations, we can use antibody-escape calculator (https://jbloomlab.github.io/SARS2-RBD-escape-calc/) informed by data from @yunlong_cao
New variant has at least as much antigenic change relative to BA.2 as does XBB.1.5.
There is P1143L at beginning of S2 stem helix
In our spike deep mutational scanning (https://cell.com/cell/fulltext/S0092-8674(23)00103-4), we found mutating P1143 improves infection by pseudotypes.
We speculated just cell-culture phenomenon, but here is mutation in virus that transmitted at least a bit.
There is deletion of V483 in RBD’s receptor-binding motif
Unpublished data from Tyler Starr and Bernadeta Dadonaite (linked in slides) indicate delV483 will moderately reduce ACE2 affinity & antibody recognition, but effect will be no bigger than some point mutations we’ve seen.
I’m going to call out three mutations that I think are especially interesting.
K356T creates a N-linked glycosylation site (at N354) in the RBD, with the resulting glycan likely to completely mask that antibody epitope.
Below is list of spike amino-acid mutations relative to BA.2, w my annotations of likely effects based on experimental data from Bernadeta Dadonaite, Tyler Starr, Yunlong Cao, and David Veesler.
One thing that is obvious is that many of these mutations cause antibody escape.
As has been noted already, this variant has lots of amino-acid mutations in spike: 33 relative to its putative ancestor BA.2. It is also very different from XBB.1.5.
This makes it an evolutionary jump comparable in size to that which originally gave rise to Omicron.