@egonw yes you're right, they are autogenerated upon deposition...
#cheminformatics nerd, professor, scientist, engineer, wife, mother, 🇦🇺 in 🇱🇺🇪🇺. FNR ATTRACT Fellow, LCSB, University of Luxembourg
#OpenScience #MassSpec #Exposome. Messages in 🇦🇺🇩🇪
@egonw you can get millions of those from PubChem by just taking the 'literature' subsection/sub selection, right?
@fresseng this is great to see!
Our proposed hackathon to improve integration of #rstats :rstats: and #Python :python: packages for #MassSpectrometry was selected for the #EuBIC2025 @EuBIC_MS developer meeting! 🥳
Looking forward to expand and improve our SpectryPy package https://bit.ly/4hQfhj1
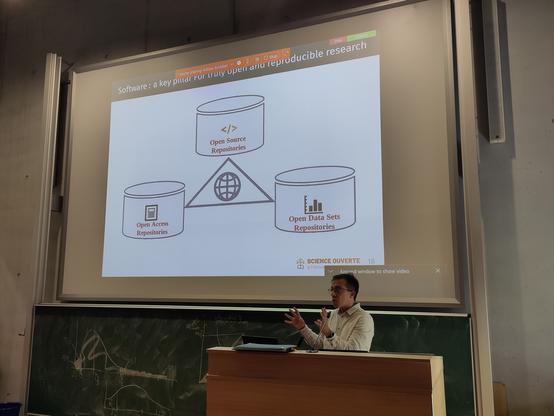
It's been an inspiring week at SSbD24 with great conversations (and great weather too). If you'd like to find out more about my take on #OpenScience and #FAIR Data for SSbD and how we try to tie it all in together you can find my slides here:
How a misinterpretation of a BMJ publication from 1996 caused (and yes, I do mean caused) the explosion in rates of peanut allergy.
In a nutshell: for decades the guidance issued in the USA and UK was the opposite of what it should have been and left a generation with preventable, life-threatening allergies.
https://news.harvard.edu/gazette/story/2024/10/excerpt-from-blind-spots-by-marty-makary/
Latest PubChemLite CSV version now available on Zenodo (Oct 2024 edition), enjoy!
Downloadable here:
https://doi.org/10.5281/zenodo.13989963
Browse here:
https://pubchemlite.lcsb.uni.lu
@fresseng nice to see!
If you gather them under a community that grows over time you can even come up with cool search URLs to find them again 🙂
https://zenodo.org/communities/lcsb-eci/records?q=presentation&l=list&p=1&s=10&sort=bestmatch
Remove the presentation from the URL and change sorting to find other goodies ... etc...
https://zenodo.org/communities/lcsb-eci/records?q=&l=list&p=1&s=10&sort=newest
It was an honour to host @fresseng from Université de Lorraine at the University of Luxembourg today to give an inspiring talk on #OpenScience this afternoon to open our International Open Access Week activities! Huge thanks to Helena Korjonen for her help organizing, we had great discussions in the lecture theatre and at the mixer afterwards!
Announcing Zotero 7, the biggest update in Zotero’s 18-year history
@egonw congratulations Dr Slenter!!!
today, Denise Slenter will defend her wonderful first Symphony of Metabolomics at 4pm CEST (with public stream): https://www.maastrichtuniversity.nl/events/phd-defence-denise-slenter #metabolomics
But we start the day with a mini symposium in the morning, with various speakers
Prof. @schymane is presenting here work at the "Symponies in Metabolomics" mini symposium
Safely home after a great #MetSoc2024 meeting in Osaka - it was a pleasure to see many familiar faces and meet plenty of new ones, and have our MassBank EU/JP meeting in person for once!
The icing on the cake was definitely being able to celebrate the honorary membership awarded to @sneumann in person! Congratulations once again Steffen, it's been a joy to be part of this journey with you!
These photos (all taken by me) are released as CC-BY.
@jorainer thanks for the pic!
Now @schymane with her keynote in the environmental exposures session at #MetSoc2024 🇯🇵
next week, Denise Slenter will defend her PhD thesis, and in the morning we are organizing a mini symposium With @schymane, @susancoort, Justin van der Hooft, Rachel Cavill, and Laura Steinbusch
If you want to join, you can register here: forms.gle/MsfrRomsiHSk2JPAA #metabolomics #wikipathways #chemistry
Denise Slenter will defend her PhD thesis on Monday 24 June 2024 at 16.00 hours. Her thesis has the title "Symphonies of Metabolomics: Utilizing graphs as sheet music to orchestrate biochemical interactions relevant in health and disease".
In the morning (9:00 to 12:30) a mini-symposium around the theme of her thesis is being organized with confirmed speakers: @schymane and Justin van der Hooft (more info soon)
@egonw cool!!! I wonder what that error was that affected 0 entries but still showed up as an error? Interesting that some duplicates are flagged, are they standardized to the same structure but starting from something different, or true duplicates?
@egonw it should take the SMILES in the CSV but you need to map the tags when you deposit the file if it doesn't have the PubChem specific headers. But it won't handle the extended SMILES of the polymers... or yes you go to SDF with the tags embedded like we did and then it should recognize everything.