@alexcc Same here but it's amazing what you can find when you spend four hours googling in front of the TV... I'll set up a list soon (this weekend?) and tweet/toot a call for contributors
A sidero-10 among siderophores 🦠
Postdoc at Wageningen University | Bioinformatician | Bioinorganic chemist | they/them 🏳️🌈
Twitter handle: @ZachReitzPhD
@alexcc Good idea! I'd be happy to maintain a microbiology list, but not so much if I have to compile all the info myself. I imagine the list will be quite a bit longer, particularly if it included the breadth of microbiology...
Down to help? Think we can get others?
Natural product researchers, do you first hear about conferences when the live tweets start or your PI leaves? I've started a list of upcoming events that feature work on secondary/specialized metabolites!
Please share and help me expand the list!
https://docs.google.com/spreadsheets/d/1-82Of7B0pjo3TqkCuIWDjqZHvceB_5VvhayLUeYtnSQ/edit#gid=0
AntiSMASH NRP metallophore prediction has only been out for three days and it already needs an update 🙃
Congratulations to Qihao Wu et al on finding a new class of metallophores! I'm looking forward to a full read and exploring that BGC!
https://pubs.acs.org/doi/10.1021/jacs.2c06410#.Y6H5AxGDwAc.twitter
@danudwary FYI, antiSMASH 7 requires python 3.9+, that was a problem I encountered.
PS: I added antiSMASH detection for three other families of metallophores: aminopolycarboxylic acids, methanobactins, and opine-like metallophores.
You'll also notice that "siderophore" clusters are now listed as "NRPS-independent-siderophore", so that the label matches the name of the biosynthetic pathway rather than the (assumed!) biological function
@danudwary Yes it is! The newest github version has it as well.
So what's next?
Well as for me, I'm looking for my next step! I've had a fantastic postdoc with @marnixmedema, but it will soon be time for me to move on. I'm looking for a position, possibly remote, that will allow me to return to my home in sunny Santa Barbara, California. If you have any leads, please let me know!
And as for metallophore discovery? We already have plans for an expanded full paper, so stay tuned!
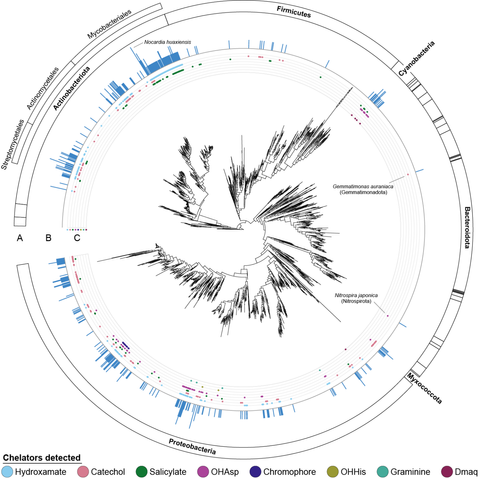
Mapping the BGCs to a species phylogeny from GTDB allowed us to see who's making (detectable) NRP metallophores. We found BGCs in 60% of Myxobacteria, 41% of Cyanobacteria, 35% of Actinobacteria, 18% of Proteobacteria, and just 3.3% of Firmicutes. Few were found outside these phyla.
Where can we find biosynthetic novelty? Cyanobacteria! We found them to be severely understudied. Nearly all BGCs were distinct from each other and from the two (!) known cyanobacterial NRP metallophores.
I applied the rules to 15k bacterial genomes to generate the first global census of NRP metallophore biosynthesis in bacteria (but certainly not the last!)
BiG-SCAPE networking showed that many BGCs are unlike anything found in MIBiG, and nearly 500 were only found in a single species! We also identified a couple of chelator combinations not found in any known BGCs
NRP metallophores have chelating substructures rarely found in other natural products, and genes encoding chelator biosyntheses are often in the BGC alongside the NRPS. I developed a series of HMMs to detect those chelator pathways.
Using this (not-so-)simple strategy, we could identify 78% of NRP metallophore BGCs with just a 3% false positive rate.
How do I know? Expert examination is still the best way to find metallophore BGCs, so I manually annotated 758 NRPS BGCs as a gold standard!
I'm pleased to announce that antiSMASH can now predict if an NRPS BGC produces a metallophore!
Check out our new preprint for all the details:
https://www.biorxiv.org/content/10.1101/2022.12.14.519525v1
I tested the new algorithm on 15k bacterial genomes. NRP metallophores make up an astounding 25% of all NRPS BGCs! This thread will have some more highlights.
Test it out on the antiSMASH website and let me know what you think!
https://antismash.secondarymetabolites.org/#!/start
(You may have to clear your cache for "Enable antiSMASH beta" to appear)
@alexcc @danudwary
'Write an argument against this statement, in 500 characters or less: "by next year we'll all be arguing with bots that are this good at arguing"'
"AI technology is not advanced enough to engage in effective arguing. It lacks the critical thinking and emotional intelligence required for effective communication and persuasion. Humans are still necessary for effective arguing."
Instead of wondering why it thinks PKSs are like tetris, you can ask!
ChatGPT is so fun
(RT me:)
Check out @ButlerLabUCSB's "In a Nutshell" article, just published in @FEBS_Letters!
https://febs.onlinelibrary.wiley.com/doi/10.1002/1873-3468.14539
In 2016, we dreamed up three hypothetical siderophores, and I set out to find them using genome mining. We've now isolated them all! (1/n)
Full thread:
https://twitter.com/ZachReitzPhD/status/1598700136036605953
@luispedro Thanks! Sounds good to me
If you're mid-tier on a 80 author paper, how do you list it on your CV?
#academicchatter
RT me
I added 60+ metallophore BGCs to MIBiG v3.0, bringing the total over 100, but there are many more I've missed! If your favorite BGC isn't in MIBiG, please let me know and I'll get it added.