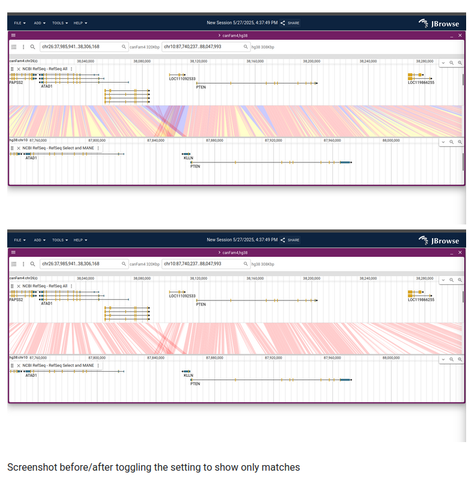
JBrowse v3.5.0 released https://jbrowse.org/jb2/blog/2025/05/27/v3.5.0-release/
This month's Alliance webinar is 'JBrowse2 at the Alliance', presented by Jeff De Pons with co-host Anne Kwitek, on Thursday May 15 at noon EDT. Please preregister by Wednesday May 14 (by midnight EDT please) to get the Zoom link https://forms.gle/GzMnmwK23SzPxbQw7
You can find the schedule of upcoming Alliance Office Hours, as well as links to the recordings of previous webinars on the Alliance Event Calendar https://www.alliancegenome.org/event-calendar
JBrowse v3.3.0 is released, featuring a big update: multi-sample SV visualization!
Users can now see complex patterns of structural variation from multi-sample VCF files. Users can also get detailed mouseover information for each sample, and see genotype frequencies in the sidebar
Release notes: https://jbrowse.org/jb2/blog/2025/04/18/v3.3.0-release/
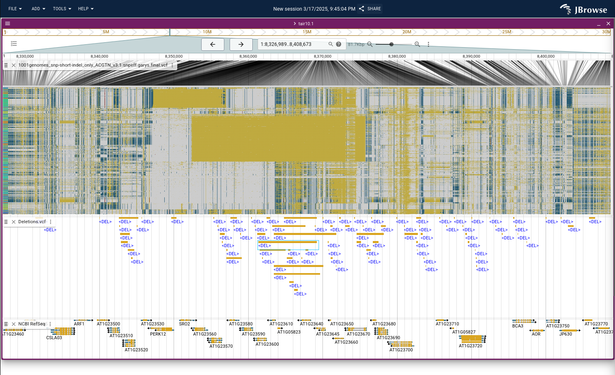
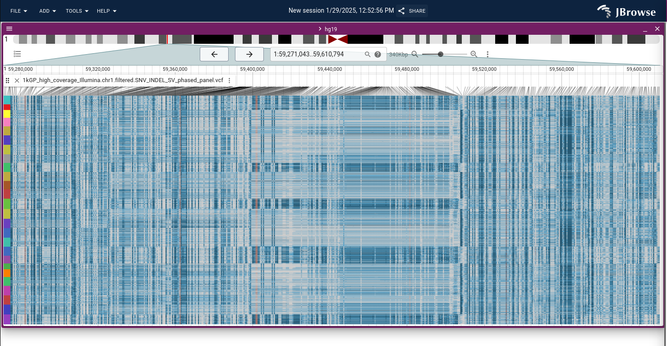
trying out the 1001 genomes data (arabidopsis) with the multi-sample variant viewer...
showing some big blocks of "no call" variants (the ./. in VCF ) in yellow...sort of indirect but these are a great 'secondary' signal for deletion SVs
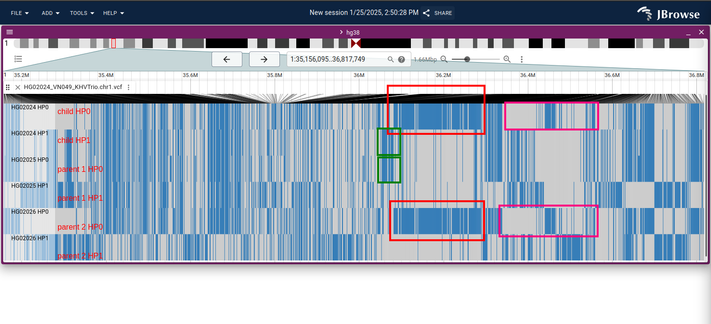
finally made a little breakthrough on finding "phased segments" from a phased VCF...had to twiddle with the hap-ibd settings to lower the defaults to output much smaller blocks. these are fragmented, but do correspond to valid blocks from what i can tell
picture shows the correspondence between hap-ibd output and the raw VCF data with a trio VCF (6 rows, each row being a haplotype of the sample)
Apollo 2.8.0 released with an important security patch
JBrowse v3.0.2 released https://jbrowse.org/jb2/blog/2025/02/13/v3.0.2-release/
I know it's a bit late, but we finally got around to writing our 2024 year in review! https://jbrowse.org/jb2/blog/2025/02/13/yearinreview/
Let us know what you think!
For those of you who are on BlueSky, we also just kicked off our BlueSky account. Give us a like and a follow :) https://bsky.app/profile/jbrowse.org
JBrowse v3.0.0 released! You can keep calling it JBrowse 2, we just did the major version bump to be good semantic versioning citizens (JBrowse 2 itself has already gone from 0.0.1->1.x.x->2.x.x->now 3.x.x)
In this release we added new phased VCF and polyploid VCF visualizations and a bunch more
Release notes here https://jbrowse.org/jb2/blog/2025/01/29/v3.0.0-release/
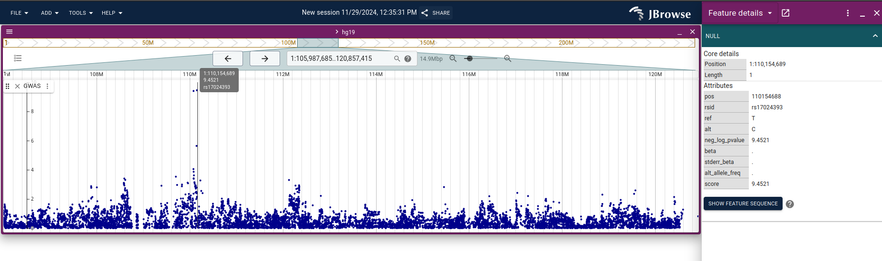
JBrowse v2.18.0 released, with new "matrix" view of variants, redesigned SV inspector, easier access to autosaved sessions, optimized GWAS view, and more! https://jbrowse.org/jb2/blog/2024/12/10/v2.18.0-release/
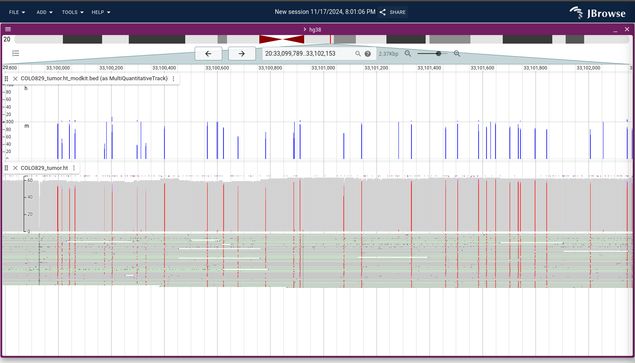
JBrowse v2.17.0 also just released with new improvements for showing base modifications and bedMethyl format, new synteny data adapter types for BLAST output, and more! https://jbrowse.org/jb2/blog/2024/11/18/v2.17.0-release/
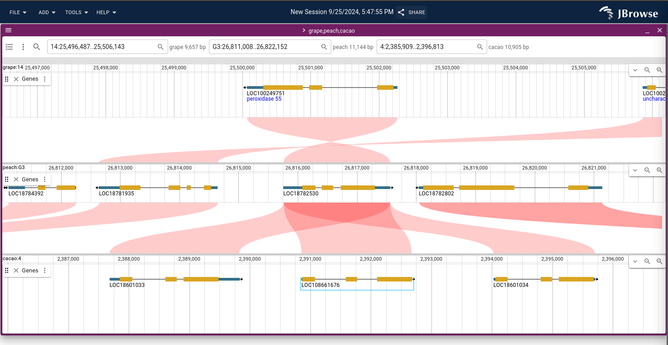
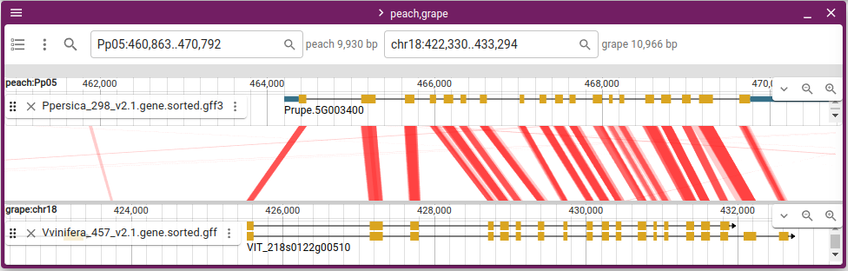
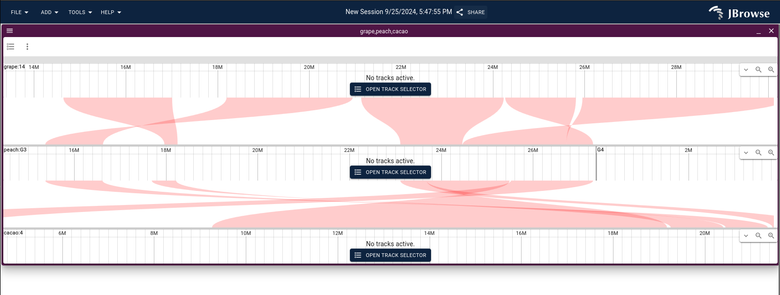
JBrowse v2.16.0 released with the ability to show multi-way synteny! https://jbrowse.org/jb2/blog/2024/10/23/v2.16.0-release/ (released a month ago, but just getting around to posting it)
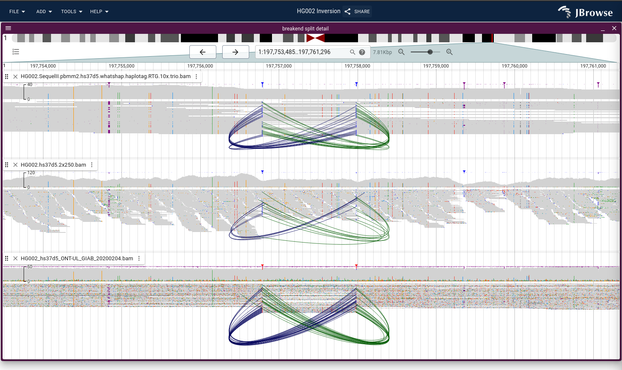
JBrowse v2.15.1 released, with some cool new features for visualizing inversions for both paired-end and split-long reads https://jbrowse.org/jb2/blog/2024/09/12/v2.15.1-release/
JBrowse v2.14.0 is now out, with some cool new features including
- Ability to group alignment track reads by specific tags
- Ability to make a FASTA file with names like e.g. NC_000001.1 to display in the UI as chr1
and more! see https://jbrowse.org/jb2/blog/2024/08/28/v2.14.0-release/ for notes
The new version also groups multiple transcripts into a gene-level feature in BigBed files automatically
In this new release, users can see genomic or feature-relative coordinates in the feature details (and pop out the side drawers into a wider screen dialog box! as pictured below)
JBrowse v2.12.0 released, with a lot of nice little features, and quality of life improvements! See https://jbrowse.org/jb2/blog/2024/06/20/v2.12.0-release/
JBrowse v2.11.2 released, with a new feature to allow users to interactively add filters to features on genome browser tracks https://jbrowse.org/jb2/blog/2024/06/03/v2.11.2-release/
We just released JBrowse v2.11.0 with new "Color by CDS" to help see which translation frame is used in the gene glyphs, and new Hi-C "color schemes" and log-transformed display https://jbrowse.org/jb2/blog/2024/04/16/v2.11.0-release/