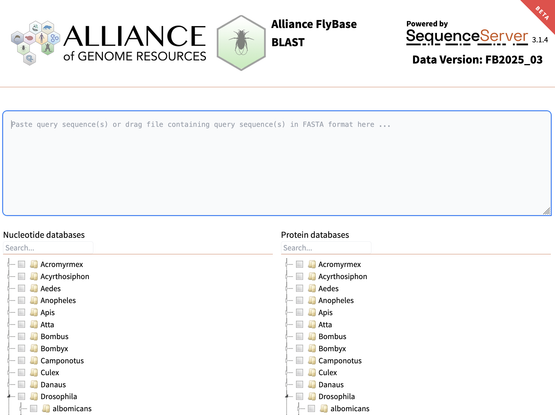
You can now search FlyBase and WormBase sequence data using the Alliance of Genome Resources BLAST server: www.alliancegenome.org/blastservice The Fly interface allows searching against many arthopods, or a search can be restricted to only Drosophila melanogaster databases.
#FlyBaseTootorial
New in FB2025_02: All Gene Group class pages,, including #SignalingPathway, #MetabolicPathway, #MacromolecularComplex and #GeneGroup reports, now have #InteractiveTables enabled.
As with other interactive tables, users can customize the 'Members' table by reordering or filtering columns, or filtering which columns to display.
#FlyBaseTootorial
and a GO-Causal Activity Model (GO-CAM https://geneontology.org/docs/gocam-overview/) that is based directly on GO annotation data.
Useful links to external resources (FlyCyc, Reactome, KEGG) and publications are provided, as well as options to export the gene list to analysis, download and orthology tools.
For more information, see this Commentary: http://flybase.org/commentaries/2024_12/MetabolicPathways.html
There's a surprise update to the #GAL4etc #QuickSearch tab!
As of today, the search results of this tab include #SplitSystemCombination hits.
#SplitGAL4 #hemidriver #FlyBaseTootorial
As part of a collaboration with the EMBL-EBI Complex Portal https://www.ebi.ac.uk/complexportal/home many fly complexes have been added to this resource and 194 Macromolecular Complex Reports have corresponding entries.
#FlyBaseTootorial #MacromolecularComplexes #GeneGroups
Please check out this Commentary about the #GeneToolKit https://flybase.org/commentaries/2024_06/GeneToolkit.html
#FlyBaseTootorial
The combination pages have been added to hitlists, so if you search for a regulatory region that drives expression of one of the hemidrivers in a combination (e.g. R72B05, VT064569, ple) you'll see the relevant combination(s) in the hitlist.
These new report pages will get improvements during the coming months, including images of expression patterns when available.
#Split_GAL4
#hemidriver
#FlyBaseTootorial
You can also find the Transgenic Product class on Allele and Transgenic Construct reports.
This release brings an update to #OrthoDB in FlyBase; we've worked with the OrthoDB https://www.orthodb.org/ team to implement a new API call that improves the accuracy of OrthoDB-derived #orthologs shown in FlyBase.
We use OrthoDB for ortholog calls of Dmel genes in many organisms (other than those represented in the FlyBase implementation of DIOPT), including other Drosophila species & arthropods.
@ensembl #FlyBaseTootorial
Click on the SO term in the “Transgenic product class” field of an Allele Report get a list of all transgenes encoding a particular type of gene product.
On the Term Report (located in the #Vocabularies tool http://flybase.org/vocabularies), you can click on the Alleles button to get a hit list of all of all the matching transgenic alleles.
#FlyBaseTootorial #TransgenicProductClass #SequenceOntology
Did you know that FlyBase has a Papers with Technical Advances page in our External Resources? These papers have been suggested by authors as including a technical advance, new type of reagent, or resource likely to be useful for other researchers, and you can find it here: https://wiki.flybase.org/wiki/FlyBase:Papers_with_technical_advances. We update this resource every month with papers that authors have submitted by flagging during the Fast-Track Your Paper process. #FlyBaseTootorial #Technical_Advances