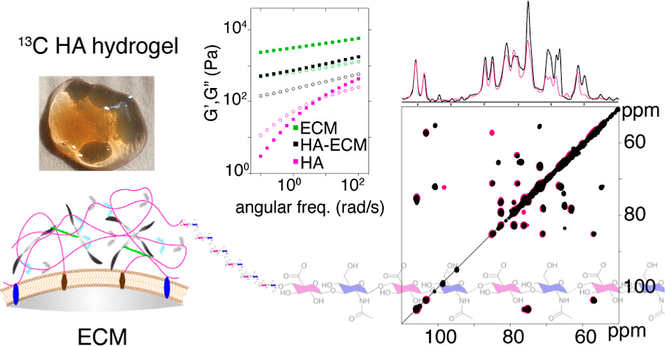
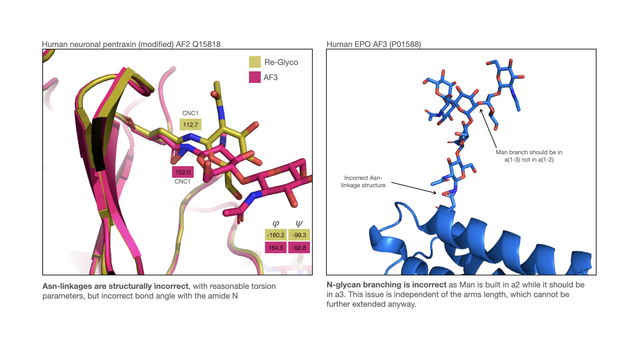
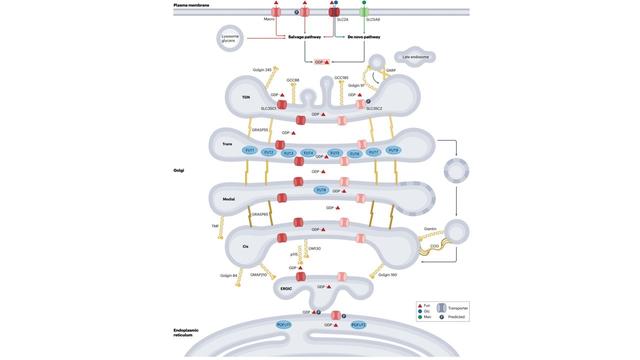
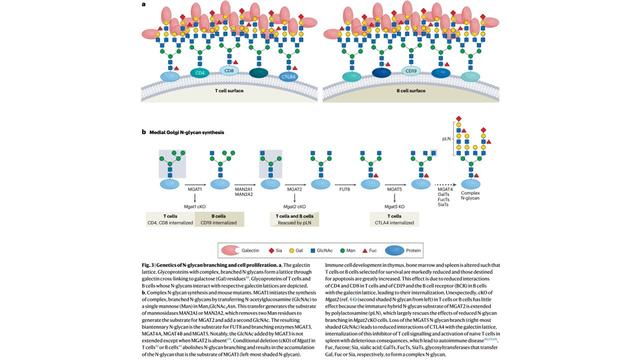
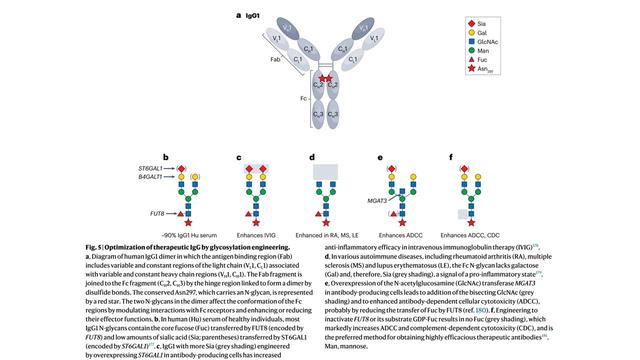
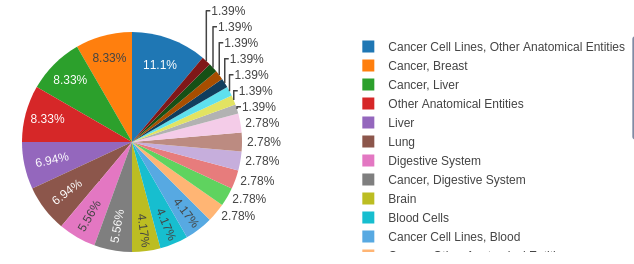
4/6 GlycoShape (https://glycoshape.org) is an OA database and toolbox that allows you to restore the 3D structure of glycoproteins with a fast and accurate algorithm named Re-Glyco.
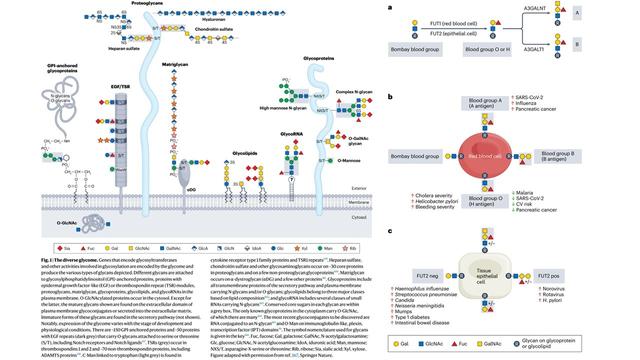
The database counts > 550 glycans to date, largely from the human glycome with examples from all species. For an overview check out the figure below ⬇️