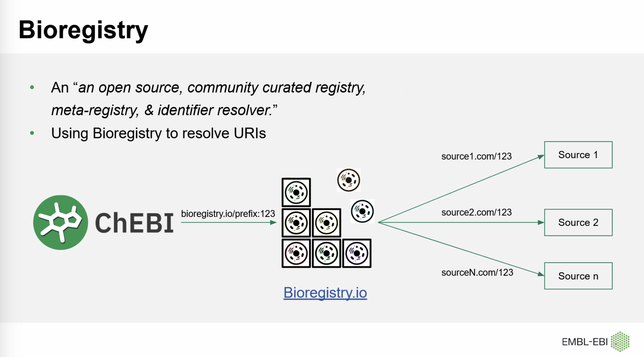
At the ChEBI 2.0 workshop, Muhammad Arsalan is presenting how ChEBI is using the Bioregistry to standardize its cross-references, generate URLs on their front-end, and more
#chebi
BridgeDb Java 3.0.27 is released: https://github.com/bridgedb/BridgeDb/releases/tag/release_3.0.27
"This update release brings in BridgeDb Datasources 20240725, a new organism (Solanum tuberosum), and various small stability improvements."
BridgeDb Java is the library that implements a Java API to access identifier mappings (e.g. from #ChEBI to #Wikidata, or from Ensembl to UniProt) from local files (there is API support for online services too, but we need help maintaining this code).
I've published a detailed article about #ChatGPT hallucination minimization using predefined #SPARQL-based query templates scoped to Linked Open Data (LOD) Cloud Knowledge Graphs such as #UniProtKB, #RHEA, #chEBI, and #OMIM.
Read: https://www.linkedin.com/pulse/reducing-hallucinations-chatgpt-using-predefined-query-idehen-4qxje/ .
#HowTo #UseCase #VirtuosoRDBMS #SemanticWeb #LinkedData #KnowledgeGraph #AI #GenAI
Remodeling other groups data is attractive but in the long run a waste of time.
Working together to remodel with the other group however is great.
Which is why I am looking forward to #CHEBI 2.0 in the ontology layer.
Of course the same query with just a changed graph name and less whitespace works at https://sparql.rhea-db.org as well. Because both #ontobee and #rhea decided not to remodel #ChEBI (no matter how much we might have wanted to). This is real #interoperability
The thing I really like about #sparql is that it is web native. I can always just send my colleagues a link to a query result. Even on databases outside of my institute, e.g. at ontobee checking for #ChEBI terms with the word mixture in their description but without a "is_a"/rdfs:subClassOf relation to CHEBI:60004 "Mixture" https://sparql.hegroup.org/sparql?default-graph-uri=&query=PREFIX+CHEBI%3A%3Chttp%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FCHEBI_%3E%0D%0ASELECT+%3Fchebi+%3Flabel+%3Fcomment%0D%0AWHERE+%7B%0D%0AGRAPH+%3Chttp%3A%2F%2Fpurl.obolibrary.org%2Fobo%2Fmerged%2FCHEBI%3E++%7B%0D%0A++++%3Fchebi+%3Chttp%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FIAO_0000115%3E+%3Fcomment+.%0D%0A++++FILTER%28CONTAINS%28%3Fcomment%2C+%22mixture%22%29%29%0D%0A++++OPTIONAL+%7B%0D%0A+++++++%3Fchebi+rdfs%3AsubClassOf+CHEBI%3A60004+.+%0D%0A++++++BIND%28true+AS+%3FisMixture%29%0D%0A++++%7D%0D%0A++++FILTER%28%21BOUND%28%3FisMixture%29%29%0D%0A++++FILTER%28%3Fchebi+%21%3D+CHEBI%3A60004%29%0D%0A++++%3Fchebi+rdfs%3Alabel+%3Flabel+.%0D%0A++%7D%0D%0A%7D%0D%0A++++++++&format=text%2Fhtml&timeout=0&debug=on
Applications session: Talk
3. Elisabeth Coudert talks about Annotation of biologically relevant ligands - such as substrates of enzymes, cofactors, activators, inhibitors as well as their binding site - in UniProt using ChEBI