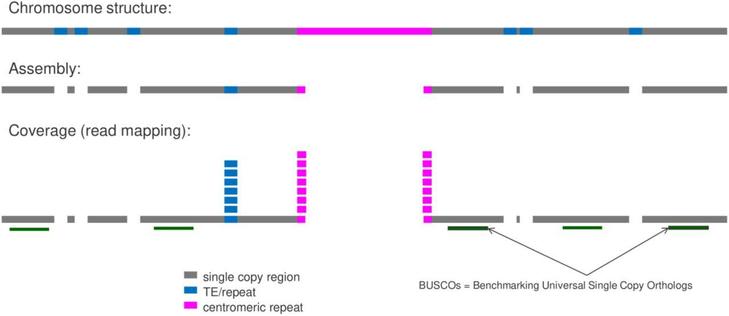
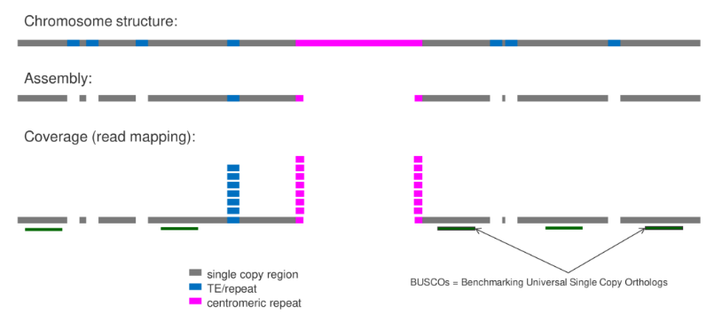
Excited to share the publication of MGSE, our new approach to genome size estimation, in BMC Genomics:
Mapping-based genome size estimation
https://doi.org/10.1186/s12864-025-11640-8
#Genomics #Bioinformatics
@PuckerLab
Molecular Plant Sciences at Uni Bonn https://www.mps.uni-bonn.de
Just wrapped up my talk at the University of Mainz on "Exploring the Specialized Metabolism of Plants with Long Read Sequencing" 🌿🔬 Great to share how nanopore sequencing helps decode complex plant pathways with ecological and biomedical potential. Thanks to the organizers and all who joined the discussion! #PlantScience #Genomics #LongReadSequencing #SpecializedMetabolism #UniversityOfMainz @PuckerLab
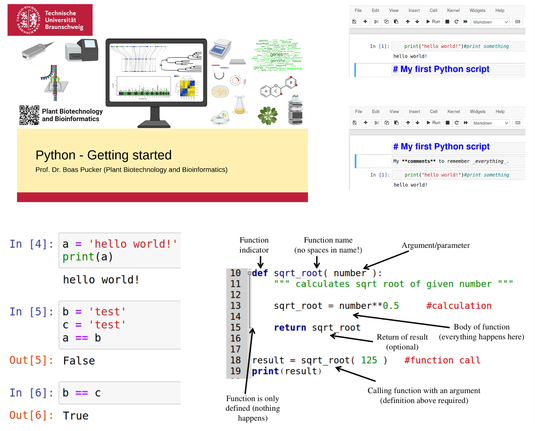
🎓 Our #Python course concludes with student project presentations covering topics like genomic feature extraction, sequence analysis, and large-scale data wrangling.
👉 See the course materials on GitHub: https://github.com/bpucker/PyBo
#OpenScience #Bioinformatics #PythonProgramming
@PuckerLab
Cornflower vibrant color comes from a unique complex of anthocyanins, flavones, and metal ions (Fe³⁺, Mg²⁺), stabilized by pH. This metal-anthocyanin interaction shifts the color to blue. #FlavonoidFriday: https://lnk.tu-bs.de/9Zz2uG
(📸 https://doi.org/10.1101/2025.01.31.635922)
We are using plant genomics & transcriptomics to understand the genetics and the evolution of the specialized metabolism of plants. Many long read sequencing projects (#ONT) are going on in the lab. We also like reusing public data sets to answer biological questions. #Python is the dominant script language in the group. We use these scripts to perform analyses beyond the potential of existing tools. Check out our website and get in touch if you would like to join: https://www.izmb.uni-bonn.de/en/molecular-plant-sciences/contact
We are using plant genomics & transcriptomics to understand the genetics and the evolution of the specialized metabolism of plants. Many long read sequencing projects (#ONT) are going on in the lab. We also like reusing public data sets to answer biological questions. #Python is the dominant script language in the group. We use these scripts to perform analyses beyond the potential of existing tools. Check out our website and get in touch if you would like to join: https://www.izmb.uni-bonn.de/en/molecular-plant-sciences/contact
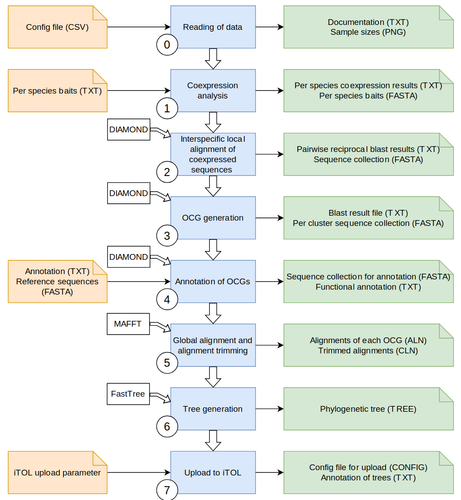
Excited to share our latest preprint about CoExpPhylo:
https://doi.org/10.1101/2025.04.03.647051.
Not long ago, Nele excelled at a #Python course for life scientists and acquired crucial skills for this work
#Bioinformatics #DataScience
@PuckerLab
Excited to share that I’ve started a new position as Professor for Molecular Plant Sciences at the University of Bonn! 🌱
@PuckerLab has officially moved, and the team is growing. Looking forward to exciting research and new collaborations ahead!
🔗 https://www.mps.uni-bonn.de
Our updated Victoria genome preprint now includes an 'Additional file 0.' Bioinformaticians, we thought you might appreciate this—what do you think?
#OpenScience #Bioinformatics #Preprints
https://doi.org/10.1101/2024.06.15.599162
@PuckerLab
While the Botanical Garden is quiet in winter, here is a stunning shot of the giant water lily, Victoria cruziana! 🌿 Curious about the molecular secrets behind its vibrant pigmentation? Dive into the research: https://doi.org/10.1101/2024.06.15.599162
#BotanicalBeauty #PlantScience #MolecularBiology
@PuckerLab @tubraunschweig
🚀 Ready to master #Python?
Today, we are diving into file handling! 📝 Learn how to:
✔️ Read data from a file
✔️ Write results to an output file
🔗 Check out the slides: https://github.com/bpucker/teaching
#Coding #TechTutorial #LearnPython
@PuckerLab @tubraunschweig
A new cohort of highly motivated life science students embarks on an exciting journey to explore the power of #Python and #Bioinformatics
@PuckerLab @tubraunschweig
Sweet news this winter! 🍫❄️ We have sequenced a cacao genome - just in time for the cozy season:
https://doi.org/10.1101/2024.11.23.624982
#ChocolateScience #WinterWonders #Genomics
@PuckerLab @tubraunschweig
The color difference between magenta and white flowers in Digitalis purpurea is linked to a large insertion in the DFR gene of white flowering plants, likely making DFR non-functional and blocking anthocyanin biosynthesis:
https://doi.org/10.1101/2024.02.14.580303
#PlantSci #Pigmentation
@PuckerLab
Excited to share the release of our Tropaeolum majus genome sequence and annotation:
https://doi.org/10.1101/2024.11.01.621629
#Genomics #LongReadSequencing
🚀 Exciting news for genome enthusiasts! 🎉 Our latest MGSE update now supports long reads and contains new data analyses for mapping-based genome size estimation. Curious about the details? Check it out: https://doi.org/10.1101/607390
#Genomics #Bioinformatics #Python
@PuckerLab
Presenting the research of our group in front of the new TU Braunschweig biology students at Burg Warberg #PlantSci
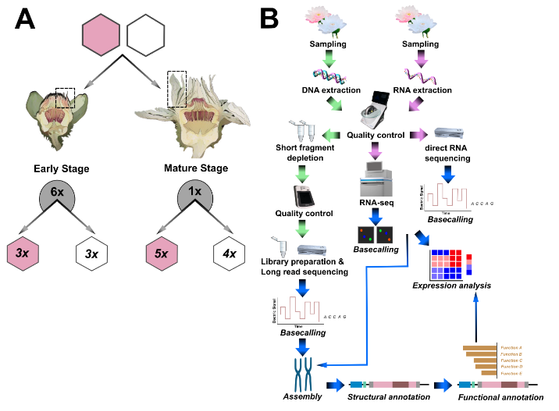
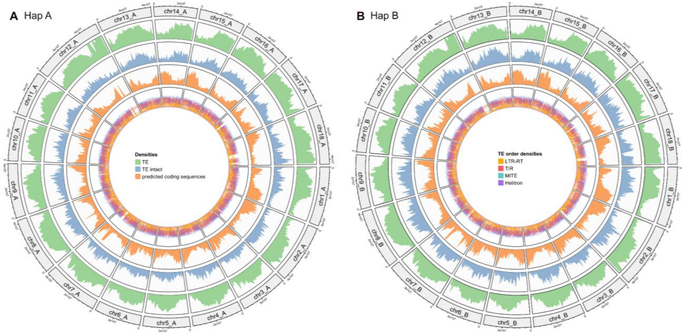
Exciting news for cassava lovers! 🥳 We have just released the phased genome sequence and annotation for the virus-resistant cultivar COL40. Key finding: a large region of repetitive elements on chromosome 12
🔗 https://doi.org/10.1101/2024.09.30.615795
#Genomics #Cassava #Research
@PuckerLab @tubraunschweig
Excited to announce the discovery of a withanolide biosynthetic gene cluster! A huge achievement from our collaboration with the Franke Lab. Explore the findings here:
https://doi.org/10.1101/2024.09.27.614867
#ComparativeGenomics #Biochemistry
@tubraunschweig @PuckerLab @unihannover
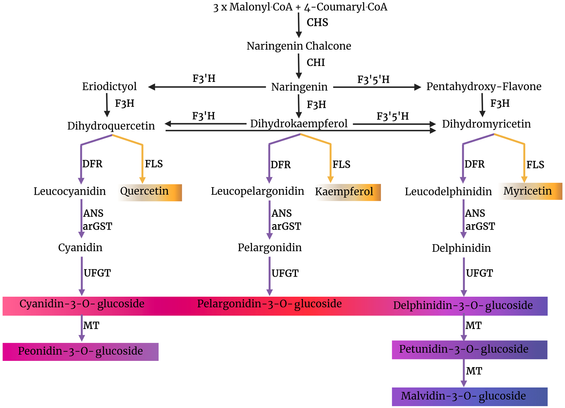
Thrilled to announce the publication of Nancy's work in PLO SONE:
Mechanisms mitigating competition between FLS and DFR
https://doi.org/10.1371/journal.pone.0305837
#PlantScience #OpenAccess #BigData
@PuckerLab