The SIB course "Gene expression made useful easily: tools and database of Bgee" is now available. I you want hands-on experience with Bgee check out our tutorial "The Bgee suite: leveraging standardized and comparable transcriptomics data across animals" at #BC2. https://youtube.com/playlist?list=PLbJBK0jNmCGJc3_20tO0P9t86bnaojhgJ&si=PjBm2cOaHS-4yL3w https://bc2.ch/tutorials-workshops @SIB @marcrr @fbastian
https://www.bgee.org Database of gene expression at @SIB and @dee_unil. #RNAseq #singlecell #biocuration #bioinformatics
The programme for the #bc2basel tutorials & workshops is out! Including our tutorial T10 - The Bgee suite: leveraging standardized and comparable transcriptomics data across animals. https://bc2.ch/tutorials-workshops https://bsky.app/profile/sib.swiss/post/3llofhltjoc2z @SIB @marcrr @fbastian
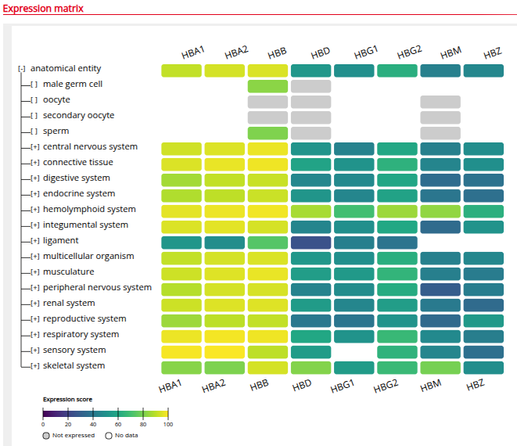
We are excited to release our gene expression heatmap visualization tool. Add genes and identify which organs or cell types the genes are highly or lowly expressed in. Try it out here: https://www.bgee.org/search/expression-matrix
@SIB @marcrr @fbastian
We had a great time at our workshop on FAIRification of single-cell RNA-Seq data and sharing the goals of #scFAIR at #biocuration2025 @biocurator
@bgeedb was used to design stem-cell based vaccines 💉against #cancer in mouse models, opening a door to a potential application with humans.
🔎 This is an example of the impact that #bioinformatics databases and tools developed by SIB groups have. Researchers use them to make sense of their data and tackle essential societal challenges.
👉 Learn more: https://www.sib.swiss/services/open-software-and-databases/impact#designing-stem-cell-based-vaccines-against-cancer
Bgee has been featured this week by the Global Biodata Coalition! Bgee is a database for retrieval and comparison of gene expression patterns across multiple animal species.
Learn more here: https://www.linkedin.com/pulse/bgee-global-biodata-coalition-ttyee
SWAT4HCLS 2024 conference proceedings are published, including Rangel et al "SPARQL Generation: an analysis on fine-tuning OpenLLaMA for Question Answering over a Life Science Knowledge Graph" which shows how Bgee's open data and integration with Wikidata can be used to train LLMs for complex queries. https://ceur-ws.org/Vol-3890/paper-4.pdf @SIB @marcrr @swat4hcls @fbastian
The SIB course “Gene expression made useful easily: tools and database of Bgee” is now available to view. This course covers the curated and pre-analyzed gene expression data in Bgee, how to detect active genes from bulk or single-cell RNA-Seq, and the various tools available to access Bgee gene expression data. Check it out! https://buff.ly/4j3Zhus
@SIB @marcrr @fbastian
📣 Don't miss the deadline! Make sure to submit your proposal of tutorial or workshop for the [BC]² Basel Computational Biology Conference before time runs out.
📅 Deadline: 14 January 2025
👉 Submit a proposal: https://tinyurl.com/2vxmsf7m
💡 Under the theme 'Bioinformatics meets AI: shaping the future of data-driven biology', the 2025 [BC]² Conference will bring together over 500 international scientists from academia, industry, and healthcare.
Bgee in 2024! Single-cell integrated with bulk RNAseq and other technologies, 52 species, and new programmatic and web access.
https://buff.ly/3BzV9Bn
@SIB @marcrr @fbastian
It's great when PubPeer allows dialogue with researchers to improve single-cell annotation 😀
https://pubpeer.com/publications/35733A35B275A626817933DE054229#
In this exciting new preprint, Warwick Vesztrocy et al use Bgee to show that "the most diverged orthologues display the greatest change in gene expression, thus suggesting that they have a greater functional divergence"
https://www.biorxiv.org/content/10.1101/2024.10.29.620890v1
@dessimoz, @go, @SIB @marcrr @fbastian @dee_unil
Left, phylogeny cartoon showing divergence between paralogs (symmetric: same branch lengths, asymmetric: different branch lengths for paralogs), and boxplots showing higher expression similarity for symmetric than assymetric paralog pairs. Right, phylogeny cartoon showing divergence of paralogs to outgroup, and graph showing higher expression similarity for slower evolving paralogs than for fast evolving ones.
Bioinformaticians of the world, it’s your time to shine! The SIB Bioinformatics Awards are now open to entries. These awards promote excellence, diversity and innovation in the field. Submit your best work and share with colleagues.
https://www.sib.swiss/bioinformatics-awards?mtm_campaign=Mastodon&mtm_source=social&mtm_medium=organic
#SIBawards #ComputationalBiology
@dessimoz @mariedangles @P_Palagi @rmwaterhouse @elixir_europe @marcrr
Delighted to represent @SIB at the AI-Bioscience Collaborative Summit in Washington DC, organized by the US Department of State & others.
Great to see a growing recognition of curated biodata’s critical role in AI breakthroughs, exemplified by resources like PDB and UniProt enabling AlphaFold.
Now, let’s focus on securing sustainable funding for these invaluable resources to continue advancing AI and bioscience innovation.
Bgee is proud to contribute open gene expression data to Wikidata. Happy birthday @wikidata!
You can leverage this data directly through Wikidata, or find it under RNA expression pattern in the info box of every gene page in Wikipedia.
https://www.wikidata.org/wiki/Q21163221
Coming to #biocuration2025? Join our workshop scFAIR: FAIRification of single-cell data https://www.stowers.org/events/biocuration2025 @SIB @biocurator @marcrr @fbastian
#scRNAseq #FAIR #biocuration #bgee #FAIRdata #FAIRification #scFAIR #SingleCellAnalysis https://sc-fair.org/
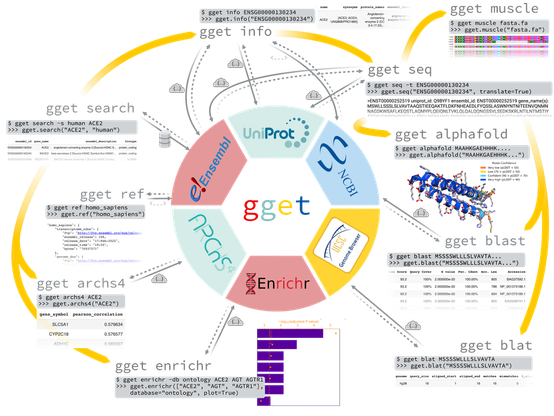
Do you know gget by the Pachter lab? You should! It now includes efficient querying of Bgee in Python. Get high quality curated gene expression data directly in Python or command line. https://pachterlab.github.io/gget/en/bgee.html #RNAseq #biocuration #Python #scRNAseq @SIB @marcrr @fbastian @dee_unil
#bgee #Bioinformatics #RNASequencing #SingleCellRNASeq #Genomics #Transcriptomics #GeneExpression #ComputationalBiology #SingleCellAnalysis #GeneExpressionAnalysis
I like when I'm following a gene-centric science talk, I check the gene in @bgeedb, and the relevant expression pattern is on top.
E.g. a talk on the role of FAM161A in retinal dystrophy, and top expression is in pigmented layer of retina in human (interestingly, Affymetrix array still providing relevant information despite all the RNAseq) and in retinal neural layer of mouse (RNAseq but also in situ hybridization, data integration FTW):
https://www.bgee.org/gene/ENSG00000170264
https://www.bgee.org/gene/ENSMUSG00000049811
Happy 30th anniversary to the Mouse Genome Database https://informatics.jax.org/, which had its first release in June 1994!
Read more about this very first release here https://www.informatics.jax.org/mgihome/news/whatsnew.shtml#jun1_2024