#YIL (Yesterday I learned) this #bioinformatics newbie gotcha: In original CIGAR strings, still widely used, 'M' denotes both matches and mismatches, while in a newer version, mismatches are separated into 'X'es 😳
Bioinformatician in Clinical Microbiology. Bioinformatics, Systems thinking, Open Science, Reproducibility, Workflow tools. Husband & Dad. Writing at https://bionics.it & https://livingsystems.substack.com
@natematias I find brave.search.com very good. I especially like the ability to get an AI-based answer based on the top results (so less flaky than a standard ChatGPT answer or similar).
Otherwise, have you tried https://devdocs.io ? I tend to have it open in a tab at all times for quick doc lookups in a number of languages.
I wrote a thing:
To do your best work you need to work like a sculptor
(How total focus, immersion and immediate feedback lets you create optimally)
https://open.substack.com/pub/livingsystems/p/to-do-your-best-work-you-need-to
Ask HN: How do you use AI/LLMs to accelerate your own learning?
https://news.ycombinator.com/item?id=42439552
🧬 Are you a #bioinformatician with some Swedish skills & interest in #microbiology looking for a job in #genomics ? 🦠
Come work with Sofia, Lili, me & co at Clinical #Microbiology at Karolinska Uni Hospital in Stockholm!
Flexible role and great people!
https://regionstockholm.varbi.com/what:job/jobID:779571/
📜New post📜
1,5 years ago I pivoted into genomics from pharmaceutical bioinformatics.
Pulling off such a pivot besides a full-time job is not easy though so I'm here sharing my experiences and recommendations for anyone else wanting to do the switch! https://livesys.se/posts/learning-genomics-bioinformatics-in-2025/
I need help! 🙏 In 2023, I compiled a list of conferences and now I would like help getting it prepared for 2024-2025. 📆 Please add/remove/edit here! https://lskatz.github.io/posts/2024/09/16/call-for-conferences.html
Do you work in #bioinformatics, love #conda & #bioconda, but didn't know how to start adding (or debugging) your/someone else's software to it?
Here's a three-part (opinionated) guide for the #microbifinie blog on just that, based on my own experiences!
Adding: https://ubinfie.github.io/2024/08/16/adding-to-bioconda-quickguide.html
Updating: https://ubinfie.github.io/2024/08/16/updating-bioconda-recipe-quickguide
Debugging: https://ubinfie.github.io/2024/08/16/debugging-bioconda-build-quickguide
Many thanks to George Bouras for nudging me to formalise my notes, and @shl , Wytamma Wirth, and @pmenzel for the testing/review!
[🧵1/2]
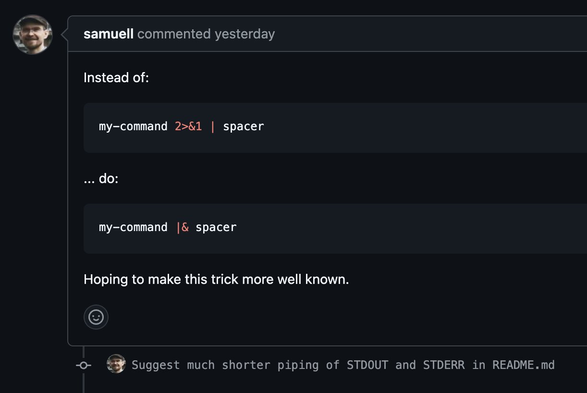
TIL |& in bash 🤯
Been working on porting my research blog (http://bionics.it) from #processwire to #Hugo
Preview available at: http://livingsys.github.io
Processwire is awesome but I feel I need to be able to edit my posts in vim & store in VCS. Wish we had the best of PW & Hugo combined 🤔
@EUCommission Hope you also rule out the offensively high fee to even enter the underground rail station at Arlanda Airport in Stockholm!
Just published in Nature Computational Science:
"Software in science is ubiquitous yet overlooked"
A comment on the many facets of software and its use in science, by a group of social scientists and computational scientists of which I am honored to be a member.
We are delighted to welcome back the Chan Zuckerberg Initiative (CZI; https://chanzuckerberg.com/science) as a Platinum sponsor of #BOSC2024!
The aim of CZI’s Open Science program is the universal and immediate open sharing of all scientific knowledge, processes, and outputs.
#snakemake 8.14 has been released. It supports setting the shell to use per rule if needed (e.g. if your analysis step runs in a container that does not have bash shell).
#sciworkflows #reproducibility
https://snakemake.github.io
btw, each time someone suggests LLMs to extract knowledge from journal articles, I cannot help but wonder why we made such a mess of knowledge dissemination in the first place
@shl awhile back i started https://cmdcolin.github.io/awesome-genome-visualization, to have an easy place to browse lots of tools in a single place. people can pick and choose!
perhaps a similar thing could be done for workflows. maybe your vision is to even further standardize workflows but that requires a larger culture change. just collecting tools in a single place is easy comparatively :)
random other note: nf-core like you mention is a great example of this, but is of course nextflow specific
@cmdcolin That page is just awesome! I have had great use of it, trying to find tools for some obscure analysis tasks. I love how you are able to get a hint of what the tools can do with a single glance.
@happykhan Well, I'm more of a CLI person, but indeed Galaxy might have some nice wrapping around common tasks, to use as a guide.
We need recipes for common bioinformatics tasks 🧬🧑🍳 New post on the blog https://bionics.it/posts/bioinformatics-recipes #bioinformatics #compbio
Tip: Stop "googling" for information. Learn how to "wiki" any subject from the wikipedia Main_Page search bar.
Bookmark https://en.wikipedia.org
Try it now. Look up anything. Then select "Further Reading" or "External Links" for the deep web we use to get from google.