Benchmarking and rebuilding bwa-mem2 with LLVM vs. GCC.
#BioConda
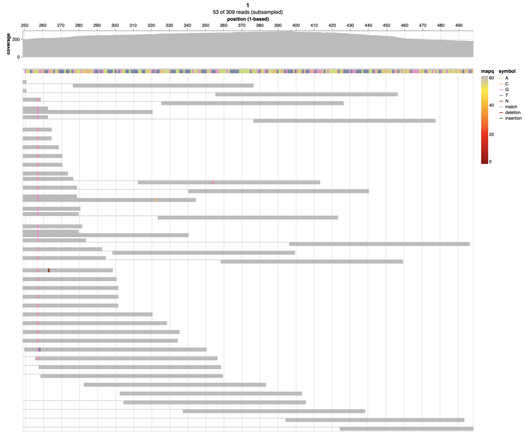
Another major new #alignoth release is now available on #bioconda ⭐ Our read alignment plots now include a coverage plot on top as well as improved visualization of overlapping read pairs! Check out our interactive demo at https://alignoth.github.io/preview.html
There is quite a backlog of open pull requests for #BioConda, with a lot of new recipes judging from the little bit of triage I just did https://github.com/bioconda/bioconda-recipes/pulls
By default, the #Seqera #Containers web interface searches the #bioconda and #condaforge channels 📦
But did you know that you can prepend your search with *any* conda channel?
Try it out: https://seqera.io/containers/
#bioconda has reached 11.111 packages. Great end of the year 2024. Thanks a lot for all contributors and the Bioconda Core team to keep the infrastructure up and running.
Genomedata 1.7.3 is now available from pip or #bioconda! Just fixes to work with newer versions of dependencies. github.com/hoffmangroup...
Release Genomedata 1.7.3 · hof...
New release for the #Snakemake #SLURM plugin to use on #HPC clusters.
A bug fix release to reliably submit and cancel jobs, where admins overwrote `sbatch` behaviour with additional output on `stderr`. See https://github.com/snakemake/snakemake-executor-plugin-slurm/releases/tag/v0.11.2 (will automatically be released on #bioconda and #pypi)
Finally resuming work on the plugin. It has been a stressful time, lately.
And if anyone from #bioconda sees this and likes it, I would be happy to chat about maybe porting it to proper docs if of interest :)
[🧵 2/2]
Do you work in #bioinformatics, love #conda & #bioconda, but didn't know how to start adding (or debugging) your/someone else's software to it?
Here's a three-part (opinionated) guide for the #microbifinie blog on just that, based on my own experiences!
Adding: https://ubinfie.github.io/2024/08/16/adding-to-bioconda-quickguide.html
Updating: https://ubinfie.github.io/2024/08/16/updating-bioconda-recipe-quickguide
Debugging: https://ubinfie.github.io/2024/08/16/debugging-bioconda-build-quickguide
Many thanks to George Bouras for nudging me to formalise my notes, and @shl , Wytamma Wirth, and @pmenzel for the testing/review!
[🧵1/2]
Lately, we’ve seen some concern and confusion among our users, so we want to clear things up. In this blog post, we’re breaking down the #conda ecosystem: the tools, the channels (like #conda-forge, #bioconda), how to get started, and how multi-stakeholder governance ensures the ecosystem remains reliable and accessible for everyone. 👉 https://conda.org/blog/2024-08-14-conda-ecosystem-explained
This was a collaborative effort from across conda ecosystem, and we hope it’s helpful for the whole community. 🤗
#bioconda #usegalaxy @elixir_europe all my favourites in one slide :-)
59 new tools in Galaxy for Exposome research!
CompareM2 is published on Bioconda! Enjoy!
For all of your prokaryotic genomes-to-report needs.
Nice - my tool's test suite passed first time on Apple M3 ARM silicon 🎉
The only catch is no #BioConda support, meaning I've skipped a few soft dependencies. And I had to locally compile a key dependency not yet ready via conda-forge.
@stackprotector @vaurora I needed to add to a #bioconda contributor's PR today, so tried this out with the gh cli.
Turns out that checking out the PR with `gh pr checkout PRNUM` does indeed set up the branch's pushRemote etc config so that pushing the branch pushes to the right fork without needing it set up as a remote. (And later deleting the branch tidies up .git/config.)
Very handy, and I may start using this instead of my home-grown equivalent.
@dryak ...and for the record:
Yes, you can #bioinformatics on a #SteamDeck -- miniforge and #bioconda work perfectly.
(not pictured above. That one was the upcoming website for V-pipe)
#Snakemake has recently reached 1 million downloads on #Bioconda! https://snakemake.github.io
#sciworkflows #reproducibility
@pjacock I assumed “they're busy” was going to be a reply to my other rant, about #bioconda being unresponsive. 🤣
I saw that biopython accidental report. Sure, they're busy, but by doing no analysis on the incoming report and just spraying it upstream in various directions, the result is more work for everyone — including themselves. Which makes everyone busier…
We passed the 100.000 software containers milestone! 🎉
The #usegalaxy project is now hosting 101,393 singularity containers for you on https://depot.galaxyproject.org/singularity/ and via CMFS. And 101,393 Docker containers are hosted by quay.io and @redhat.
#biocontainers rocks!
Thanks to de.NBI @elixir_europe and the amazing #bioconda community.