so, maybe you are wondering how we're doing with the 1 million IUPAC names? https://chem-bla-ics.linkedchemistry.info/2025/03/08/iupac-names.html https://chem-bla-ics.linkedchemistry.info/2025/04/27/one-million-iupac-names-2-the-100-thousand-milestone.html
Well, last week the 200k milestone was passed. But we're also running out of #openaccess articles from Europe PMC.
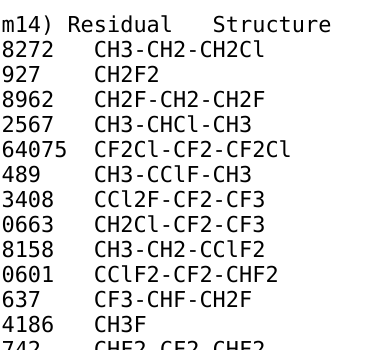
By doing some simple string replacements (e.g. methyl/ethyl/propyl) we can grow the size easily to beyond 1 million! That is not bad, with three months of running scripts in the background.
But first #ICCS2025 and teaching grading first.