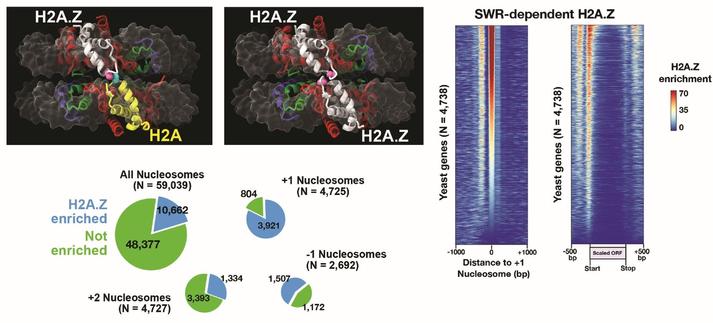
How is the H2A.Z #histone variant targeted to specific sites in the genome by the SWR #chromatin remodeler? This study shows that DNA sequences contribute to SWR specificity and that SWR’s H2A.Z insertion activity is stimulated by polyA tracts in #nucleosomes @PLOSBiology https://plos.io/4kggWyD
#Nucleosomes
Single #nucleosomes tracked in live cells during #cell_division using super-resolution #microscopy.
https://phys.org/news/2024-08-nucleosomes-tracked-cells-cell-division.html
Organization Of DNA In Cell
#ArchaeaDNA, #DNA, #Histone, #Nucleosomes, #Supercoil #Genetics All living cells contain DNA in the form of double helix, its organization differs among cells in the three domains…. Medical Microbiology & Recombinant DNA Technology (RDT) Labs | Read More -
https://micrordt.wordpress.com/2024/04/27/organization-of-dna-in-cell/
To find #nucleosomes, chromatin remodelers slide and hop along #DNA, and their direction of approach affects the direction that nucleosomes slide in. https://elifesciences.org/articles/96836?utm_source=mastodon&utm_medium=social&utm_campaign=organic_insights
Biologists discover the secrets of how #gene traits are passed on.
#epigenetics #inheritance #nucleosomes #DNA
https://phys.org/news/2024-03-biologists-secrets-gene-traits.html
"Is X-ray crystallography dead?" some asked when #cryoEM got momentum, then when #AlphaFold arrived. Depends on what you work on. For #nucleosomes it certainly looks like it is dead: all structures with nucleosomes released in 2023 were cryoEM structures!
On this #ThrowbackThursday we go back to our Episode #58 where we talked with Efrat Shema from the Weizmann Institute of Science about her work on Single Molecule Imaging of #chromatin, and the analysis of #nucleosomes circulating in plasma. #epigenetics
Listen here: https://www.activemotif.com/podcasts-efrat-shema
Validation of high-speed atomic force #microscopy as a tool for measuring the dynamics of individual #nucleosomes in vitro. https://elifesciences.org/articles/86709?utm_source=mastodon&utm_medium=social&utm_campaign=organic
Changes in adenoviral #chromatin organization precede early gene activation upon infection
Harald Wodrich, Gernot Laengst and colleagues show that host #nucleosomes replace #adenoviral genome packaging before viral genes are transcribed.
https://www.embopress.org/doi/full/10.15252/embj.2023114162
New microscopy assay by Sue Biggins and collaborators #fredhutch enables direct observation of coordinated de novo assembly of native #CENPA #nucleosomes driven by several protein cofactors and the AT run content of #centromeric DNA
#RefereedPreprint c/o @ReviewCommons
https://www.embopress.org/doi/10.15252/embj.2023114534
How the #epigenetic landscape modulates pioneer #transcription factor binding.
#nucleosomes #Yamanaka #histones
https://phys.org/news/2023-05-epigenetic-landscape-modulates-transcription-factor.html
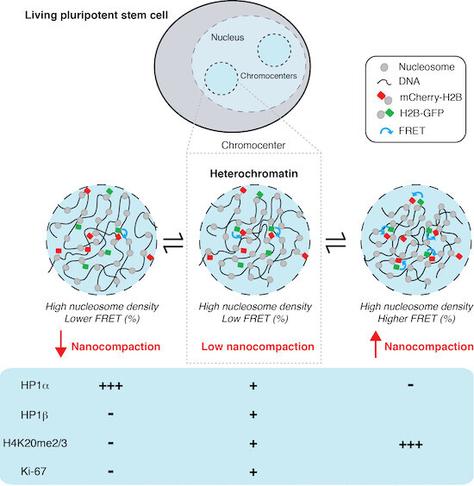
Surprises from #FLIM-#FRET studies of nanometer-range distances between #nucleosomes in living, non-fixed cells:
Claire Dupont, Robert Feil, David Lleres et al show that constitutive #heterochromatin is in fact less compacted than bulk #chromatin
How the 'master #regulators' of cells make #DNA accessible for gene #expression.
https://phys.org/news/2023-04-master-cells-dna-accessible-gene.html
#DNA #packaging supports #cell division, finds study.
#mitosis #replication #ORC #nucleosomes
https://phys.org/news/2023-04-dna-packaging-cell-division.html
The molecular basis for recognition of #CENPA #nucleosomes by ‘Mis18 licensing factor component #KNL2:
Tatsuo Fukagawa and coworkers present #cryoEM structure with chicken KNL2 and insight into its dynamic localization regulation during the #cellcycle
https://www.embopress.org/doi/10.15252/embj.2022111965