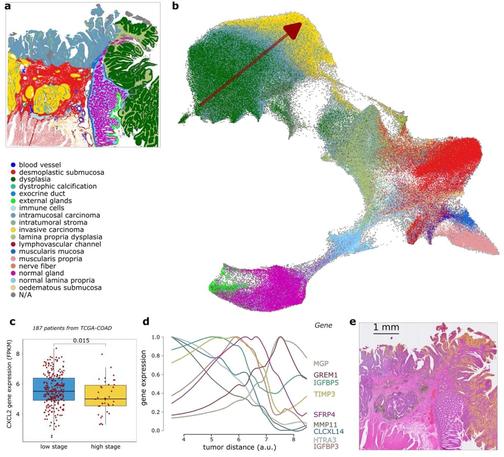
KODAMA goes spatial
https://doi.org/10.1101/2025.05.28.656544
#spatialtranscriptomics #visium #visiumhd
#SpatialTranscriptomics
Want a sneak peek or to try it on your own data? Try EnrichMap https://enrichmap.readthedocs.io/en/latest/
#SpatialTranscriptomics #ComputationalBiology #EnrichMap #spatialomics
🧬🔬🧪 We are seeking a Scientist to push the limits of spatial transcriptomics in zebrafish! Come join us!
#MolBio #SpatialTranscriptomics #DevelopmentalBiology #ScienceJobs 🧫🐟🔍
job-boards.greenhouse.io/chanzuckerbe...
Scientist, Spatial Molecular C...
Decoding cell–cell communication using spatial transcriptomics
#spatialtranscriptomics #omics #ccc
https://www.nature.com/articles/s41576-025-00824-3
📣 Listen up: you only have a few more days to apply for #EMBOSpatialOmics!
Would you like to get a comprehensive introduction to the various methods of spatial-omics and in particular those of multiplex immunofluorescence and spatial transcriptomics?
Then apply for this hands-on EMBO Practical Course by 10 March ✍️
🌍 EMBL Heidelberg
📅 15 – 20 June 2025
💻 https://s.embl.org/iso25-01-ma
#spatialtranscriptomics #microscopy #imageanalysis #spatialbiology #spatialomics
Spatiotemporal single-cell roadmap of human skin wound healing
#scRNA-seq
#spatialtranscriptomics
#visium
#RNAScope
https://www.cell.com/cell-stem-cell/fulltext/S1934-5909(24)00412-0?dgcid=raven_jbs_aip_email
#job alert: junior/mid-career PI opportunities in cancer research @ Oxford university: https://www.ludwig.ox.ac.uk/news/vacancies-assistant-or-associate-professor-level-cancer-researchers
Areas we're looking for:
#epigenetics #chromatin #epitranscriptomics #RNA #myeloidCells #spatialTranscriptomics
Please boost for visibility! @academicchatter #hiring
Tools (#preprint from 10x Genomics).
'Here, we introduce Visium HD Spatial Gene Expression (“Visium HD”) and demonstrate its use as a discovery platform for profiling CRC in multiple patients using FFPE tissue blocks. Visium HD slides provide a dramatically increased oligonucleotide barcode density over the Visium v2 slides (11,000,000 continuous features in a 6.5 mm Visium HD capture area, compared to 5,000 features with gaps between them in a 6.5 mm Visium v2 capture area). The single cell scale resolution of Visium HD allowed us to map distinct populations of immune cells, specifically macrophages and T cells, and evaluate differential gene expression at the tumor boundary to explore the potential contribution of these immune cell populations in the TME.'
#Immunology #Immunotherapy #SpatialTranscriptomics
https://www.biorxiv.org/content/10.1101/2024.06.04.597233v1
Some really great talks and discussions today on advances in virus-livestock omics at the #Turing institute, who kindly supported the event. Talks include those by Ge Wu and Marie Di Placido (pictures) from the #Pirbright Institute.
Great to have input from the #BBSRC and #SFTC #Harttree Centre too.
Looking forward to new adventures in this area...
#livestock #virus #genomics #antibody #genome #mutation #pangenome #omics #transcriptomics #spatialtranscriptomics #fmdv
Last chance to register for our meeting "Advancing virus-livestock omics" on Thurs 28th Sept at the Alan Turing Institute.
Registration closes end of Mon 25th.
See https://www.turing.ac.uk/events/advancing-virus-livestock-omics
Register here: https://t.ly/wYA7d
Schedule below
Zoom stream possible for those from further afield
#livestock #virus #genomics #antibody #genome #mutation #pangenome #omics #transcriptomics #spatialtranscriptomics #fmdv
Marius Lange #BC2basel: increasing complexity of #SingleCell data leads to problems of temporal, spatial and spatio-temporal mapping. The solution proposed is optimal transport applications, implemented in Moscot:
https://moscot.readthedocs.io/en/latest/
https://www.biorxiv.org/content/10.1101/2023.05.11.540374v2 #SpatialTranscriptomics #scRNAseq
Excellent conclusions of Jean Fan #BC2basel on #machinelearning applied to #scRNAseq #SpatialTranscriptomics
Impressive work by Jean Fan's lab #BC2basel: aligning different spatial transcriptomics results, not only from different experiments but different protocols, e.g. MERFISH - Visium. Combining with #scRNAseq clustering and her lab's spot deconvolution allows to compare directly gene expression between these different experiments, https://www.biorxiv.org/content/10.1101/2023.04.11.534630v2.abstract #SpatialTranscriptomics
There is Minecraft. And then there is the #Visium manual fiducial alignment
Excited for participating in this #spatialtranscriptomics #Visium platform study with an expert team from @NICHD_NIH, in the first step of studying the spatial biology of #pituitary corticotroph tumors, revealing their heterogeneity.
https://www.medrxiv.org/content/10.1101/2023.08.04.23293576v1
I might as well have make a pinned post.
We published Vesalius a tool that uses Image analysis methods to analyze spatial-omics data.
A new version will be coming soon that will include multimodal analysis and alignment!
GitHub: https://patrickcnmartin.github.io/Vesalius/
MSB: https://www.embopress.org/doi/full/10.15252/msb.202211080
Computational Scientist
Icahn School of Medicine at Mount Sinai
Come join our team as a #computationalScientist: #multiOmics modeling of #myeloma, including #singleCell, #spatialTranscriptomics and #flowCytometry
See the full job description on jobRxiv: https://jobrxiv.org/job/icahn-school-of-medicine-at-mount-sinai-27778-computational-scientist-5/?feed_id=47998
#ScienceJobs #hi...
https://jobrxiv.org/job/icahn-school-of-medicine-at-mount-sinai-27778-computational-scientist-5/?feed_id=47998
Postdoctoral Researcher
Icahn School of Medicine at Mount Sinai
Come join our team as a #bioinformatics postdoc: #multiOmics modeling of #myeloma, including #singleCell, #spatialTranscriptomics and #flowCytometry
See the full job description on jobRxiv: https://jobrxiv.org/job/icahn-school-of-medicine-at-mount-sinai-27778-postdoctoral-researcher-4/?feed_id=47893
#ScienceJobs #hi...
https://jobrxiv.org/job/icahn-school-of-medicine-at-mount-sinai-27778-postdoctoral-researcher-4/?feed_id=47893
O Psoriasis, a troubling foe,
We seek to know what causes your woes.
Your lesions so hard to manage
What makes them range?
Our search will make your secrets known.
#psoriasis #genetics #spatialtranscriptomics #research #ode #poetry
Computational Scientist
Icahn School of Medicine at Mount Sinai
Come join our team as a #computationalScientist: #multiOmics modeling of #myeloma, including #singleCell, #spatialTranscriptomics and #flowCytometry
See the full job description on jobRxiv: https://jobrxiv.org/job/icahn-school-of-medicine-at-mount-sinai-27778-computational-scientist-5/?feed_id=45913
#ScienceJobs #hi...
https://jobrxiv.org/job/icahn-school-of-medicine-at-mount-sinai-27778-computational-scientist-5/?feed_id=45913