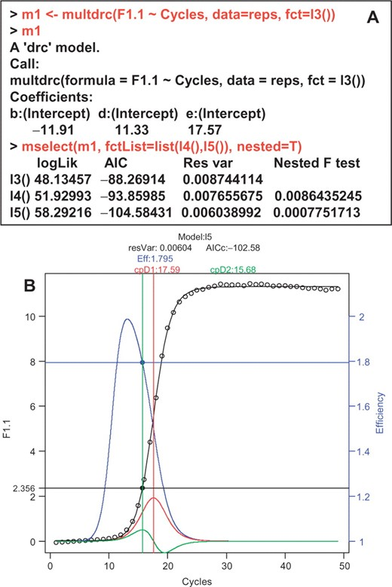
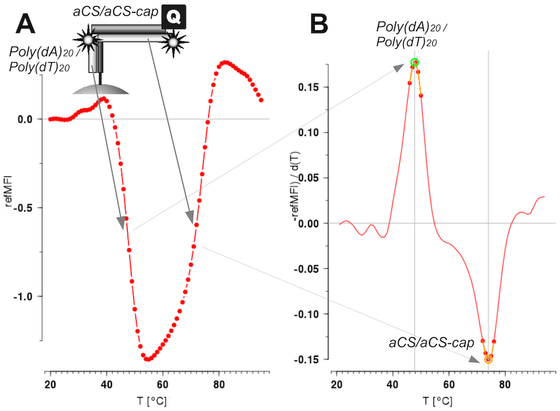
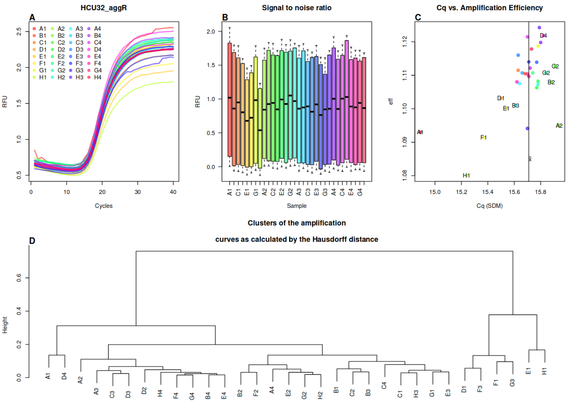
Coming soon in @Mol_Ecol
Omics to study and manage aquatic environments: a snapshot from the AquaEcOmics meeting (Evian-les-Bains, 2025)
F Rimet, C Lemonnier, B Alric, P Beja, L Bittner, J Bylemans, F Leese, R Logares, K Meissner, F Not, L Orsini, B Paix, N Rodríguez-Ezpeleta, R Siano, B Thalinger, N Tromas, V Vasselon, I Domaizon
Thanks to all co-authors! Great to work with you and to summarize this great #aquaecomics meeting #omics #metabarcoding #eDNA #metabolomics #dpcr #metagenomics