Read this preprint from Manon Berger, Lester Kwiatkowski, Laurent Bopp, and me about how if you consider iron limitation, growing seaweed doesn’t work as a CO₂ removal (CDR) technique.
Microbiologist.
I'm posting more at bsky.app/profile/acritschristoph.bsky.social than on here these days.
My American friends: I hope you join me today.
To stand up for our freedom to assemble & our freedom of speech. To reject authoritarianism & dictatorship.
To demand the government be accountable to the people - not the reverse.
Talking Climate is now a podcast! Every week, you can listen to the good news, not-so-good news, and what you can do on Spotify and Substack, narrated by the amazing Anne Cloud from Voice Over For The Planet.
Whether you're commuting, doing chores around the house, or even if you're just tired of looking at a screen, this new podcast version is for you. My hope is that having an audio version makes it easier than ever to stay informed and inspired.
We’ve already got nine episodes ready to binge — please enjoy, and let me know what you think!
Listen on Spotify: https://open.spotify.com/show/583MdhAsDvn46YcPpvOJkR?si=59acbdf0ee0b4a35
Listen on Substack: https://www.talkingclimate.ca/podcast
Coming soon to all of your favorite podcast platforms.
Climate misinformers focus on the stats that we don't predict will change, e.g., number of landfalling hurricanes.
See the post here: https://www.theclimatebrink.com/p/climate-change-is-making-hurricanes-caf
We often hear folks skeptical of climate change claim that "the climate is always changing" or "temperatures rise before CO2" or "its the sun!". In a new episode of PBS's Weathered, Jessica Tierney and I try and address these questions: https://www.youtube.com/watch?v=iP2lH2EEr9I
Prevalence and Dynamics of Genome Rearrangements in Bacteria and Archaea
https://www.biorxiv.org/content/10.1101/2024.10.04.616710v1.abstract
This special issue highlights more than a decade of progress in applying #genomics and advanced #bioinformatics to #publichealth. It includes real-world examples of how technologies are transforming #surveillance, #outbreak response, #antimicrobialresistance monitoring, and #pandemic preparedness.
The May 2025 supplemental issue of Emerging Infectious Diseases journal is now live: “Advances in Pathogen Genomics for Infectious Disease Surveillance, Control, and Prevention.”
https://wwwnc.cdc.gov/eid/articles/issue/31/13/table-of-contents
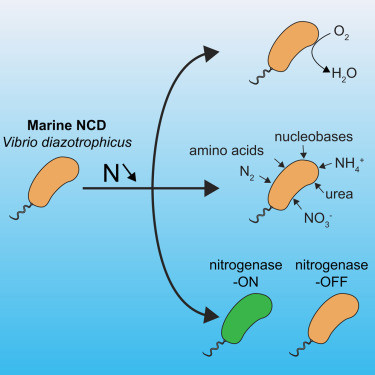
High metabolic versatility and phenotypic heterogeneity in a marine non-cyanobacterial diazotroph https://www.cell.com/current-biology/fulltext/S0960-9822(25)00568-8?rss=yes&utm_source=dlvr.it&utm_medium=mastodon
Excited to announce the start of ARTIC2! 🎉
ARTIC2 builds on the success of ARTIC aiming to make sequencing more equitable and accessible. 🤝🧬
EVE Group at Swiss TPH is a key team member - we'll be helping make sequences easier to store and share via Loculus & @pathoplexus ! 🌎
More info in the link! 👀👇🏻
https://www.birmingham.ac.uk/news/2025/ambitious-project-to-develop-low-cost-genome-sequencing-for-pathogens-known-and-unknown
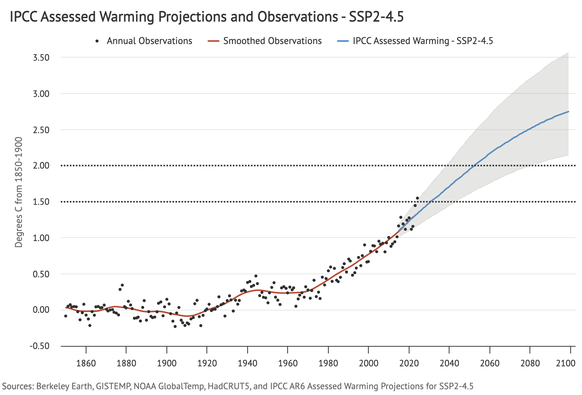
Global temperature over the past two years have been very much on the high end of the range of future warming expected in a current policy-type scenario (SSP2-4.5) in the most recent IPCC report (which, itself, expects some near-term acceleration):
@foaylward Congrats, amazing paper! Just read it, saw your name, and remembererd you were active on here!
Consider also joining us on bsky - we have a pretty vibrant microbial ecology community there (hence why I haven't checked in or posted as much here recently)
We found a really cool giant virus inside the genome of a green alga, and it has a very strange infection cycle that is hard to detect.
There are reports going back decades of viral particles spontaneously appearing in pure cultures of green algae, so this phenomenon seems quite common but mostly overlooked.
Sustained in situ protein production and release in the mammalian gut by an engineered bacteriophage
Interstitial microbial communities of coastal sediments are dominated by Nanoarchaeota https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1532193/full
Do you make core genome alignments for phylogenomics? Mona Taouk and I explored how including sites with some missing data (a soft core) can improve analysis, especially for large datasets.
https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.001346
(1/4)
We have a new preprint describing a method for #bacteriophage phylogenetics and comparative genomics.
Maybe useful for those interested in all the amazing #phages out there and their bizarre mosaic genomes
Towards a unifying phylogenomic framework for tailed phages
https://www.biorxiv.org/content/10.1101/2024.10.21.619452v1.abstract
As part of the Public Health Alliance for Genomic Epidemiology (PHA4GE) wastewater working group we’ve been working on developing a series of guidance documents for wastewater pathogen surveillance work, and these documents are now public (https://github.com/pha4ge/wastewater-guidance/tree/main)!
We intend this repository to serve as a live, open community resource that anyone is more than welcome to contribute to, and contributions can be made directly on github via a pull request.
@cidgoh and @pha4ge are developing ISO-based, modular, interoperable contextual data standards supporting global Mpox genomic surveillance and sharing. To learn more, find them here: One Health Mpox: https://github.com/cidgoh/MPox_Contextual_Data_Specification, Wastewater: https://github.com/pha4ge/Wastewater_Contextual_Data_Specification.