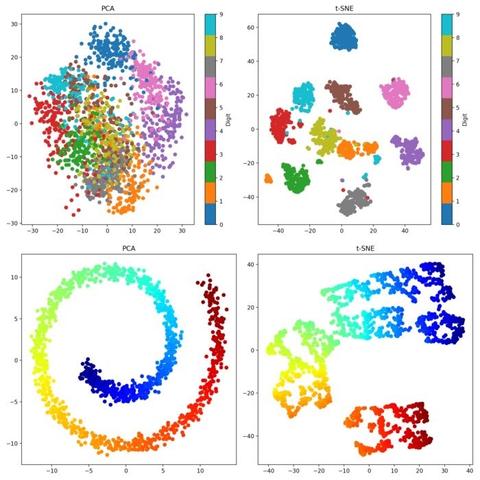
#ReleaseTuesday — New version of https://thi.ng/tsne with ~15-20% better performance[1] due to avoiding repeated internal allocations and skipping gradient updates where unnecessary...
[1] Benchmarked with multiple datasets of ~750 items, each with 192 dimensions (now ~165ms @ MBA M1, 2020)...
#ThingUmbrella #TSNE #DataViz #Visualization #ML #Cluster #TypeScript #JavaScript