Single cell RNA-sequencing (#scRNAseq) is an essential method to learn about cells in health and disease. Here we have studied "multiplets", an important source of error of scRNAseq. We find that multiplets are astonishingly frequent and hard to eliminate.
https://doi.org/10.1101/2025.06.09.658708
#scrnaseq
Pipeline release! nf-core/scnanoseq v1.2.0 - nf-core/scnanoseq v1.2.0 - Copper Rhinoceros!
Please see the changelog: https://github.com/nf-core/scnanoseq/releases/tag/1.2.0
#10xgenomics #longreadsequencing #nanopore #scrnaseq #singlecell #nfcore #openscience #nextflow #bioinformatics
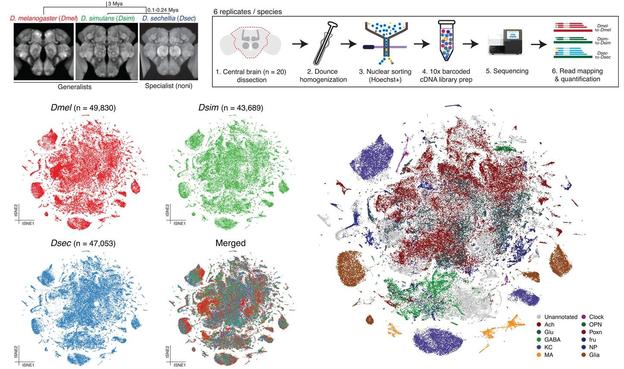
How do #brain cells change over #evolution? @bentonlab compare #scRNAseq from ecologically distinct #drosophilid species to identify changes in composition & gene expression of different cell types, revealing higher divergence in #glia than #neurons @PLOSBiology https://plos.io/4js7Rms
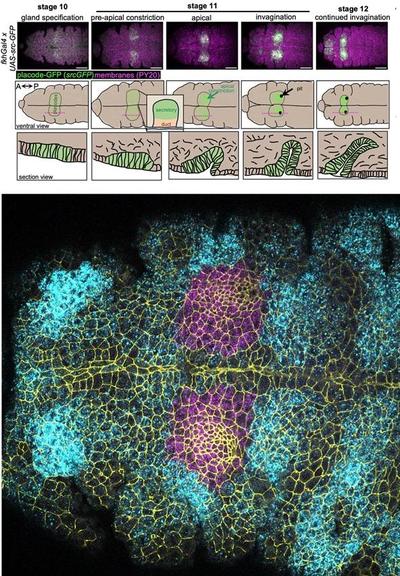
How does transcriptional patterning regulate #SalivaryGland #morphogenesis? Annabel May & @katjaroeper use #scRNAseq of early morphogenesis of the #Drosophila salivary gland placode to reveal regulation by induction & exclusion of regulatory factors @PLOSBiology https://plos.io/4cUw827
Our new preprint is now out!
Dynamic transcriptional heterogeneity in pituitary corticotrophs
https://www.biorxiv.org/content/10.1101/2025.04.04.645979v1
We analysed publicly available single-cell RNA sequencing data of pituitary gland tissue and looked at corticotrophs, cells that are central to mediate stress responses.
We identified several transcriptional states in these cells that are related to how they respond to stress. Cells are able to transition between these states and this might be helpful for them to respond to stress coming at unpredictable times.
We also highlight issues related to using scRNAseq to look at functional subpopulations of cells.
#scrnaseq #stress #physiology #cellbiology #bioinformatics #corticotrophs #pituitary
Pipeline release! nf-core/scnanoseq v1.1.0 - nf-core/scnanoseq v1.1.0 - Iron Alligator!
Please see the changelog: https://github.com/nf-core/scnanoseq/releases/tag/1.1.0
#10xgenomics #longreadsequencing #nanopore #scrnaseq #singlecell #nfcore #openscience #nextflow #bioinformatics
Uneven #hormone distribution in #plants regulates #cell_division and #growth, biologists discover.
#Brassinosteroids #signalling #cell_cycle #agriculture #scRNAseq
https://phys.org/news/2025-03-uneven-hormone-cell-division-growth.html
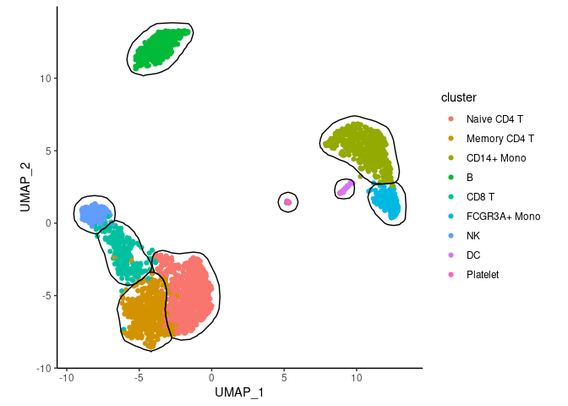
mascarade package implements a procedure to automatically generate 2D masks for clusters on single-cell dimensional reduction plots like t-SNE or UMAP https://github.com/alserglab/mascarade #rstats #scRNAseq
chatomics! How to Fine-Tune the Best Clustering Resolution for #scRNAseq Data 🎯 🧵
Finally, our paper studying synovial tissue #DC #DendtriticCells in joint health and #rheumatoidArthritis is out in Immunity!
#scRNAseq #spatialTranscriptomic
From: @ImmunityCP
https://mstdn.science/@ImmunityCP/113557582489769767
Computational toolbox from Human Cell Atlas.
#scrnaseq #humancellatlas
https://www.nature.com/articles/d41586-024-03762-y
2D embeddings for high-dimensional data, the battle rages on
https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1012403
Insbesondere bei der Interpretation von #RNA-Sequenzanalysen einzelner Zellen (#scRNAseq) sind Techniken zur Dimensionen-Reduktion wie #UMAP der neuste Schrei. Doch bilden UMAP-Plots tatsächlich die Realität ab? Es gibt Zweifel … Zur Methoden-Kontroverse: https://www.laborjournal.de/rubric/hintergrund/hg/hg_24_11_01.php
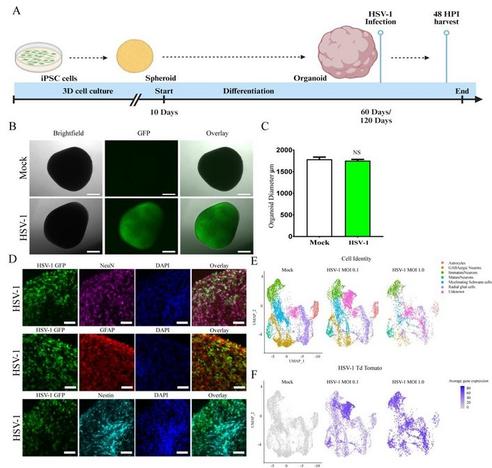
🔊 New pre-print from David Butler and team!
"HSV-1 Infection Alters MAPT Splicing and Promotes Tau Pathology in Neural Models of Alzheimer's Disease"
#organoid #tauopathy #neurodegeneration #scRNAseq #Neuracell
biorxiv | https://zurl.co/iWU1
An interesting bioRxiv preprint was shared on the 🐦 site (https://x.com/strnr/status/1844105666962579813). The paper describes a model to represent cells from large scale scRNA seq atlases using LLMs. Apart from the novelty value one of the main draws should be the ability to map any dataset with no additional data labelling, model training or fine-tuning onto the existing universal cell embedding. https://www.biorxiv.org/content/10.1101/2023.11.28.568918v2
https://github.com/snap-stanford/UCE
#scRNAseq #embedding #biology #llm
Coming to #biocuration2025? Join our workshop scFAIR: FAIRification of single-cell data https://www.stowers.org/events/biocuration2025 @SIB @biocurator @marcrr @fbastian
#scRNAseq #FAIR #biocuration #bgee #FAIRdata #FAIRification #scFAIR #SingleCellAnalysis https://sc-fair.org/
It’s been a while since I’ve had to design a #scrnaseq pipeline. What tools are folks using for optimizing clustering resolution, or is it all still done iteratively (I.e. by “feel” using bio knowledge) #bioinformatics #singlecellrnaseq #clustering #genomics
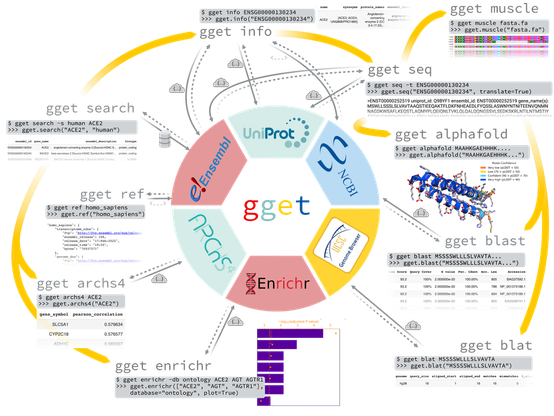
Do you know gget by the Pachter lab? You should! It now includes efficient querying of Bgee in Python. Get high quality curated gene expression data directly in Python or command line. https://pachterlab.github.io/gget/en/bgee.html #RNAseq #biocuration #Python #scRNAseq @SIB @marcrr @fbastian @dee_unil
#bgee #Bioinformatics #RNASequencing #SingleCellRNASeq #Genomics #Transcriptomics #GeneExpression #ComputationalBiology #SingleCellAnalysis #GeneExpressionAnalysis
paraCell: A novel software tool for the interactive analysis and visualization of host-parasite single cell RNA-Seq data (without knowing programming) https://biorxiv.org/content/10.1101/2024.08.29.610375v1 . Interested in the datasets? Here: http://cellatlas.mvls.gla.ac.uk
"By using time-resolved analyses of scRNA-seq data, we determined the potential transitional trajectories of tumor cells and identified the metastasis-initiating subpopulations"
https://link.springer.com/article/10.1007/s11684-024-1081-7
Reading right now. The identification of cells that initiate #metastasis are of interest, although n=2 paired primary and #BoneMarrow samples may be a bit limited.