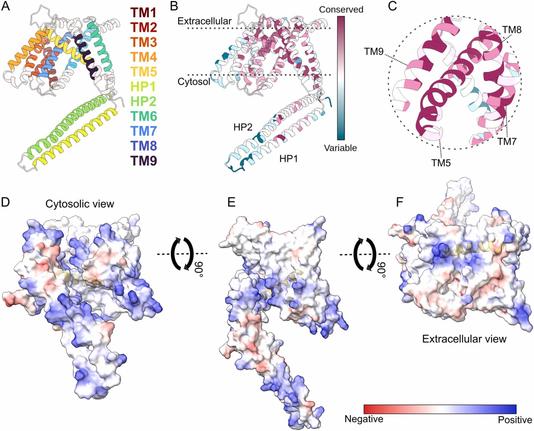
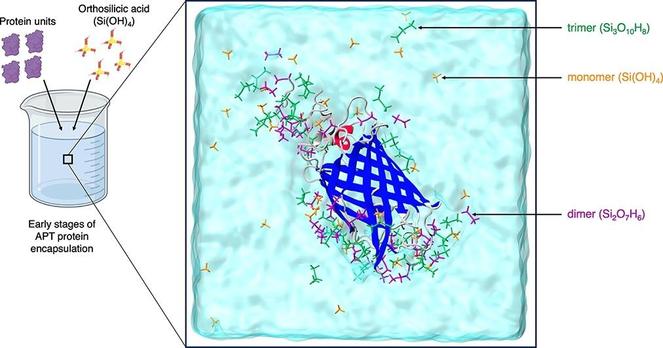
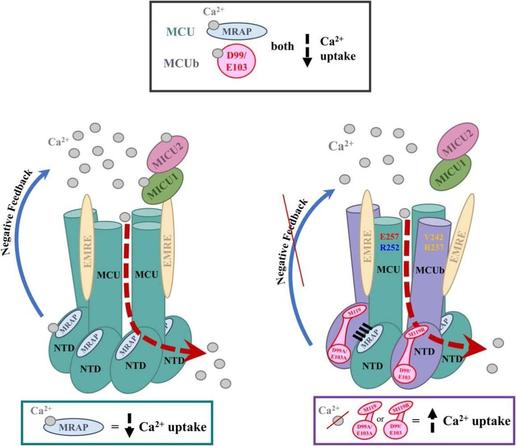
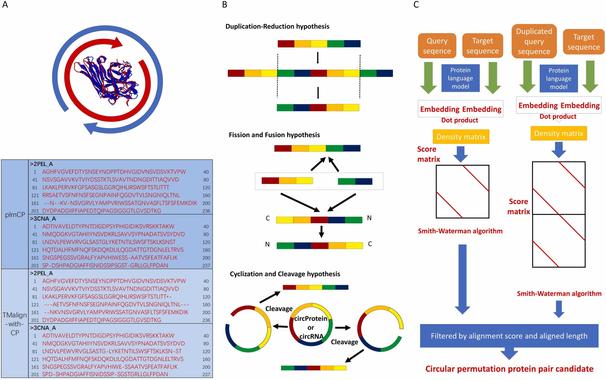
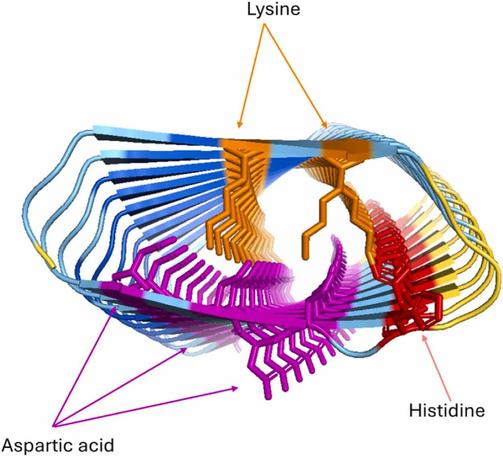
🧩 What if the diverse functions of a single protein all trace back to one structural superfamily?
🔗 Evolutionary and structural bioinformatics identifies GPR89 as a conserved member of the LIMR protein superfamily. Computational and Structural Biotechnology Journal, DOI: https://doi.org/10.1016/j.csbj.2025.11.003
📚 CSBJ: https://www.csbj.org/
#StructuralBiology #ComputationalBiology #MolecularEvolution #ProteinScience #Genomics #Proteomics #AlphaFold #MembraneProteins #Genetics #Bioinformatics