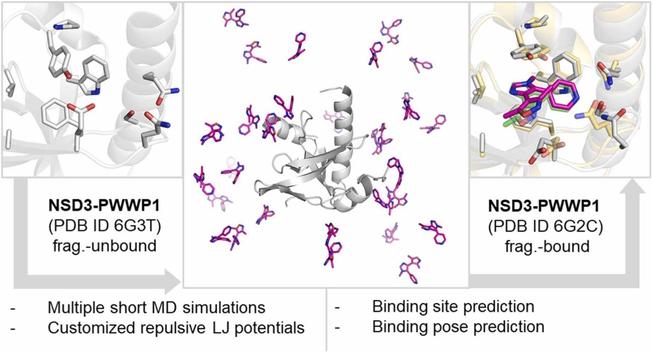
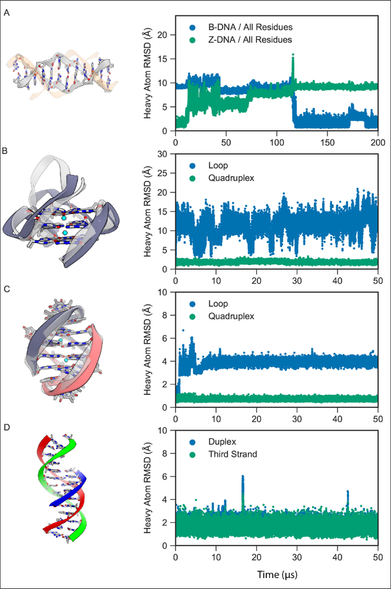
I am shamefully copying this post from Linkedin but if you happen to know any candidate, do not hesitate to reach:
It's on. To my #ComputationalChemistry network, I am calling on you. 🙇
The PM2E team in the CIMAP laboratory (www.cimap.ensicaen.fr) is looking for a #PhD hashtag#candidate. The goal of the project is to understand key parameters in #semiconductor defects stability and physics (both vacancies and color centers, see for example: https://doi.org/10.1016/j.nimb.2022.12.025) through the use of
#DFT and #MolecularModeling. The end goals of the project is concerned with application for #QuantumComputing through the making of stable #qubit. Collaboration and communication with experimentalists will be part of the project.
If you're interested in #MolecularDynamics, #MonteCarlo simulations, or DFT and quantum calculations, we are waiting for you. We are using codes such as #Lammps and #QuantumEspresso. We are looking for a candidate finishing or already having a master degree (or equivalent diploma) with training in #physics/ #MaterialsScience / #TheoreticalChemistry. Any experience in programming using #Python, #C and the #Linux environment would be considered a plus.
The position is located in Alençon (Orne) between Caen and Le Mans. Founding is assured for relevant candidates.
Link to the offer (in french, you can ask for an english version): https://www.abg.asso.fr/en/candidatOffres/show/id_offre/127985/job/simulation-atomistique-de-defauts-crees-par-irradiation-dans-les-semiconducteurs