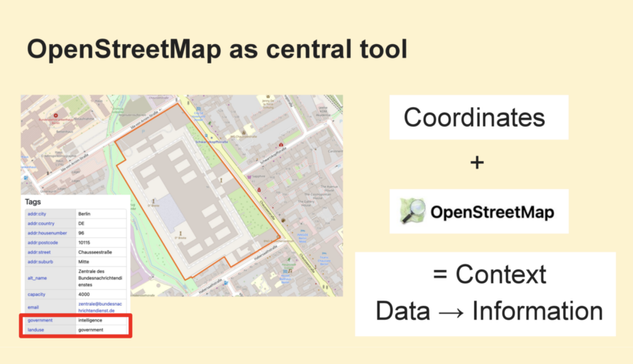
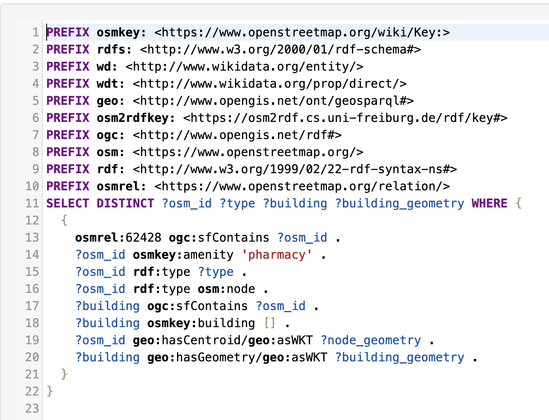
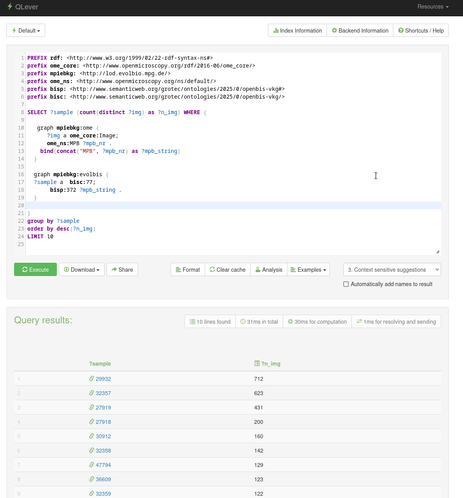
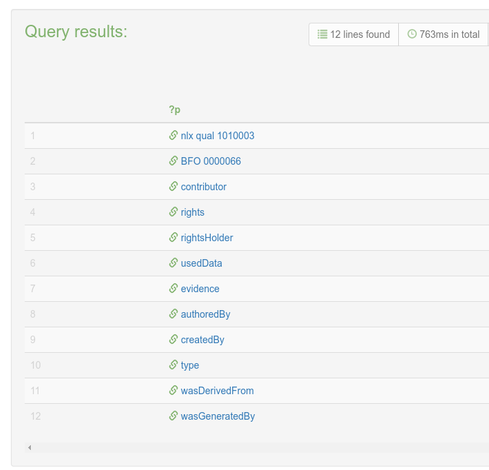
Service-Link zur heutigen @digiSberlin -Veranstaltung anlässlich des #Digitaltag2025 (vgl. https://openbiblio.social/@digiSberlin/114732577091825896)
Folgender Query listet die Online-Kataloge/Online-Sammlungen der Berliner #GLAM-Einrichtungen: https://qlever.cs.uni-freiburg.de/wikidata/jGfXMD (Query powered by #qlever . Die zwei unbegrenzten property paths zwingen blazegraph in die Knie ...)
Folien zur Veranstaltung folgen ...