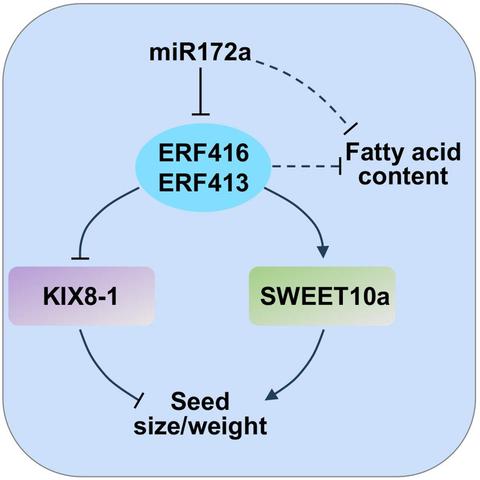
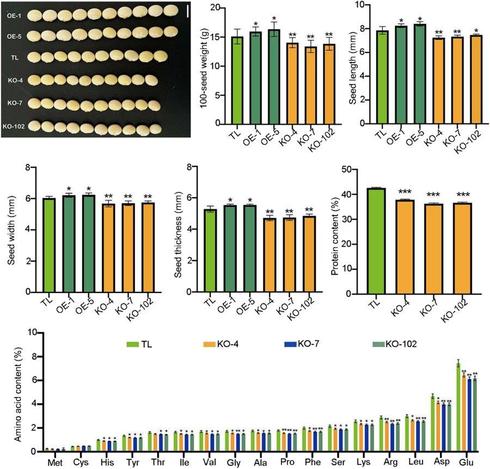
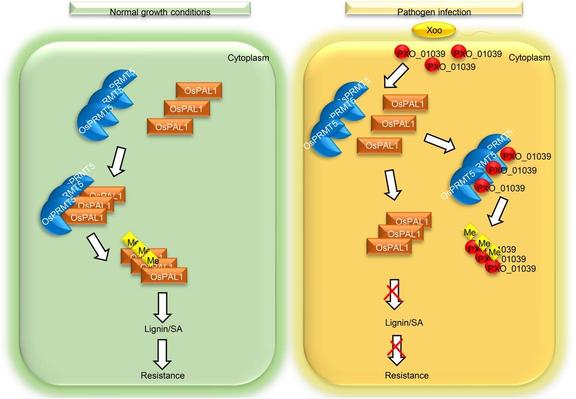
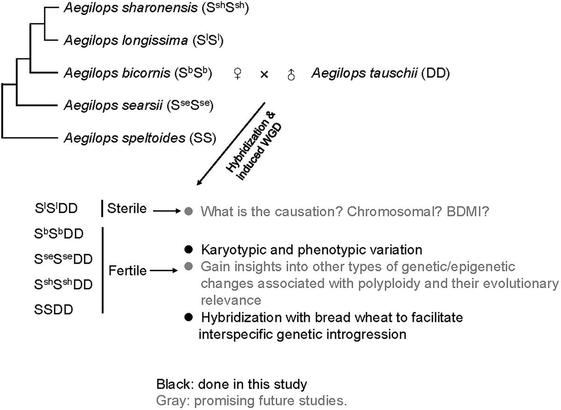
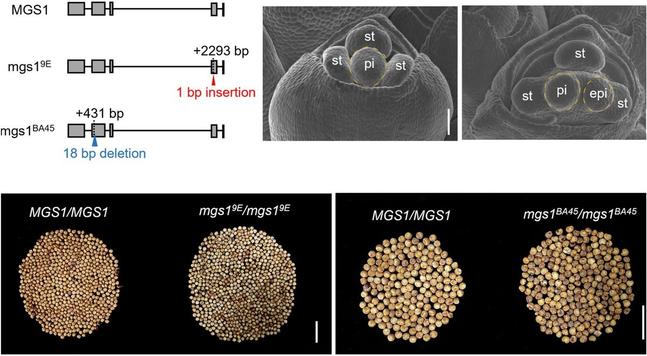
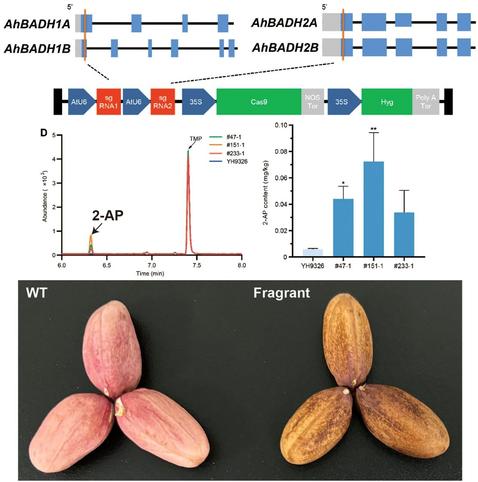
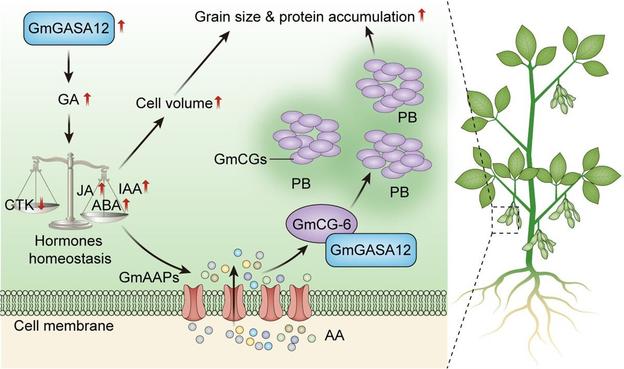
In this #OpenAccess research article, Jin et al. reveal a new module for #seed trait control in #soybean, and explore the potential use of these alleles to facilitate breeding for high-#oil and high-yield cultivars.
Read their findings here⬇️for #free!
https://doi.org/10.1111/jipb.70015