and in supp data the logo of diat.barcode ;-) #diatoms #barcoding #eDNA #molecularecology
#barcoding
Happy to share our new paper presenting a new version of diat.barcode reference #barcoding #dna #library for #diatoms #nature #rivers #water #science #ecology
Gelis, Barry-Martinet, Briand, Chonova, Cochero, Gassiole, Kahlert, Karthick, Keck, Kelly, Kochoska, Mann, Nayak, Pfannkuchen, Servulo, Trobajo, Vasselon, Vidakovic, Viollaz, Wetzel, Zimmermann, Rimet, F. (2025). https://doi.org/10.1051/limn/2025009
Genetic Diversity of Ukrainian Populations of Invasive Species of the Genus Galinsoga Assessed by ISSR-Markers - #barcoding #biologicaldiversity #geneticpolymorphism #moleculargenomics #interspecieshybridization #molecularmarkers #Ukraine #ISSR #Galinsoga - https://link.springer.com/article/10.3103/S0095452725010141
Universal #RNA #barcoding system for tracking #gene_transfer in #bacteria created.
https://phys.org/news/2025-03-universal-rna-barcoding-tracking-gene.html
Die Frage "Was krabbelt denn hier?" ist bei #Spinnen nicht leicht zu beantworten. Yannis Schoeneberg von der Trier möchte samt Kollegen per Multi-Locus-#Barcoding das „taxonomische Rennen“ gewinnen, bevor es zu spät ist … Mehr dazu von Karin Lauschke: https://www.laborjournal.de/editorials/3197.php
#dnaderived #sequences #barcoding #Cortinarius Added another several dozens of sequences to our Fungarium YSU database, which soon will be discoverable though GBIF. Hope to find collaboration through shared barcoding initiatives. Especially like these days Cortinarius spp., they are amazing.
Totally about 1K specimens in our collection have dna-derived data (attached sequences): https://www.gbif.org/dataset/d922b606-6c94-4d51-9277-36c9b03872a7
Nice presentation of Adriana Radulovici on #iBOL #DNA #dnaquaimg #barcoding @leeselab
20 years after Paul Hebert paper on DNA #barcoding who could expect this wordwide success?
Nice presentation of carbon footprint of molecular methods by O Monnier and his student Mathieu.
#footprint #metabarcoding #algae #eDNA #barcoding #carbon
Excellent paper from F Keck.
Reference barcoding libraries problems:
(i) mislabelling, (ii) sequencing errors, (iii) sequence conflict, (iv) taxonomic conflict, (v) low taxonomic resolution, (vi) missing taxa and (vii) missing intraspecific variants. They give possible consequences on the taxonomic assignment process
Navigating the seven challenges of taxonomic reference databases in metabarcoding analyses
https://doi.org/10.1111/1755-0998.13746
#metabarcoding #DNA #bioinfo #barcoding
Single degenerated #primer significantly improves #COX1 #barcoding performance in #soil #nematode profiling.
https://phys.org/news/2024-02-degenerated-primer-significantly-cox1-barcoding.html
Revisiting Israel's #freshwater #fish species list through cutting-edge #DNA #barcoding technology.
https://phys.org/news/2024-01-revisiting-israel-freshwater-fish-species.html
Visit our webpage about phytoplankton metabarcoding
https://carrtel-collection.hub.inrae.fr/barcoding-databases/phytool
#algae #phytoplankton #metabarcoding #eDNA #barcoding #INRAE #Carrtel
Visit ourwebpage about diatom metabarcoding methology
https://carrtel-collection.hub.inrae.fr/barcoding-databases/diat.barcode
#diatom #metabarcoding #eDNA #barcoding #algae #INRAE #Carrtel
RT by @Plants_EFSA: Behind the scenes, indentify #2 : @aimethods DNA Barcoding.
Black-headed leafcutter bee, Megachile circumcincta. The DNA was isolated from a leg of the bee using a liquid (lysis buffer). Means everything dissolves in it except for the chitin. Then the Co1 gene was amplified with PCR. This means that certain parts of the DNA are copied so often that they are easily detectable. Detectable for the sequencer, the next step. The 313 base pairs required for the identification are then determined here. (See colorful graphic) With the help of a worldwide database of already certain animals whose Co1 gene has been determined, the base pairs can be compared with each other.
#dnabarcoding #barcoding #dna #macro #insects #bumblebee #hummel #insekten
🐦🔗: https://nitter.cz/sagaOptics/status/1725182377717108888#m
[2023-11-16 16:01 UTC]
Nice courses that we gave to students in University of Torino with Kalman Tapolczai on diatom metabarcoding. Thank you to Francesca Bona and Elisa Falasco for the invitation
@Kalipster
#algae #eDNA #barcoding #metabarcoding #diatom #biomonitoring
Just published: "Marine invasive alien species in Europe: 9 years after the #IAS Regulation"
Proud to contribute to the #eDNA part of this review on #BioMonitoring of #marine #invasive #alien species in Europe, as part of the #GES4SEAS project. Have a look, esp. if you wondered how many #NIS species for which sequence vouchers exist for #DNA #barcoding or #metabarcoding
#biodiversity #genomics #training 🧑🏫with BGE partners perfecting techniques for HMW #DNA 🧬 extractions for #genome assembly & plates of #arthropods 🐞 for #barcoding hosted by Auth University Thessaloniki 🇬🇷
@erga_biodiv @SIB
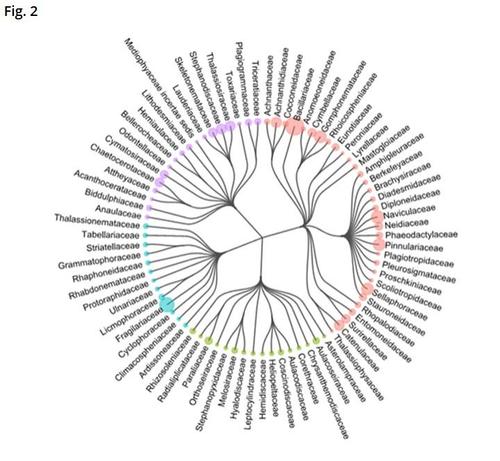
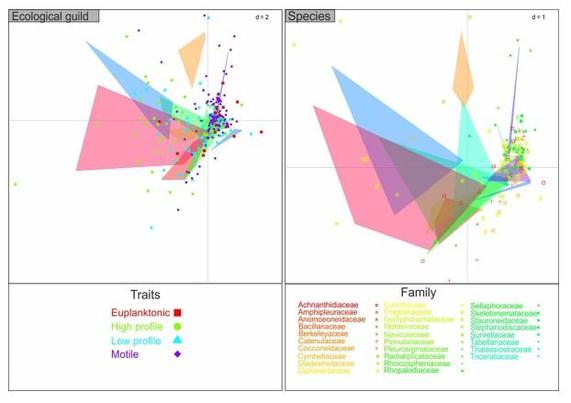
Here is our new paper about diatom metabarcoding :😋
Filling reference libraries with diatom environmental sequences: strengths and weaknesses
https://www.tandfonline.com/doi/full/10.1080/0269249X.2023.2237977
#edna #metabarcoding #diatom #barcoding #diversity #biodiversity
Here is a free read-only link to our latest paper on retention and export of planktonic fish eggs in the northeastern Gulf of Mexico #fish #barcoding #DNA #oceanography #science #USFCMS #fisheries
https://onlinelibrary.wiley.com/share/author/XTB7CTTCBYSMSBUGBNNA?target=10.1111/fog.12655
Very proud of our new paper:😀
Canino, A., C. Lemonnier, B. Alric, A. Bouchez, I. Domaizon, C. Laplace-Treyture, & F. Rimet, 2023. Which barcode to decipher freshwater microalgal assemblages? Tests on mock communities. International Journal of Limnology EDP Sciences 59: 8, 1-15. DOI: 10.1051/limn/2023008
https://www.limnology-journal.org/articles/limn/full_html/2023/01/limn230014/limn230014.html