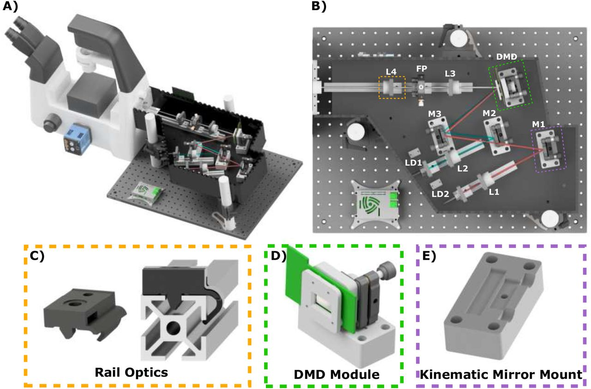
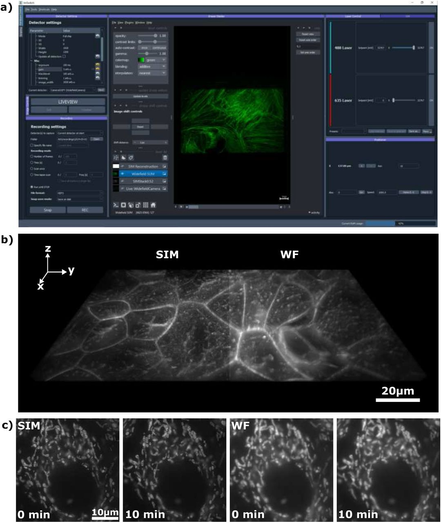
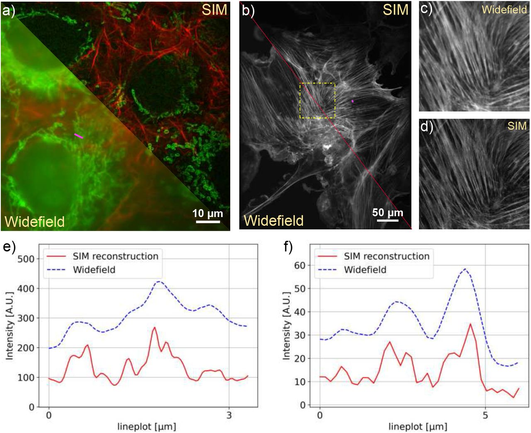
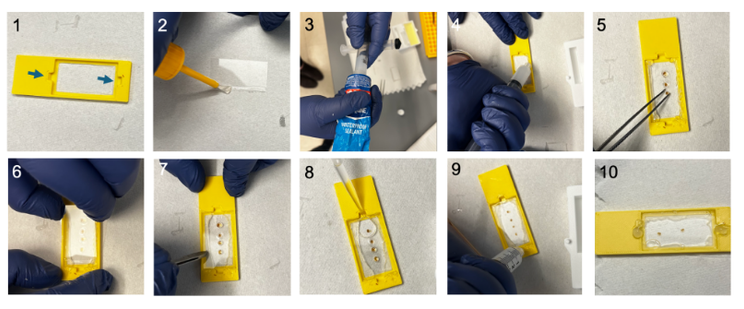
Image analysts: What is your go to method for batch converting image files from the proprietary format to a tiff file? If this should be done at all...
I’ve been using FIJI to open and re-save mine. Although this gives me another look at the image as a QC step, I am using Napari more and more and have found a tiff easier to load into Python.